产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# DF10308, RRID:AB_2840886.
展开/折叠
ARNT like protein 1 brain and muscle; Arntl; Aryl hydrocarbon receptor nuclear translocator like; Aryl hydrocarbon receptor nuclear translocator like protein 1; Aryl hydrocarbon receptor nuclear translocator-like protein 1; Basic helix loop helix PAS orphan MOP3; Basic helix loop helix PAS protein MOP3; Basic-helix-loop-helix-PAS protein MOP3; bHLH PAS protein JAP3; bHLH-PAS protein JAP3; bHLHe5; BMAL 1; BMAL1_HUMAN; BMAL1c; Brain and muscle ARNT like 1; Brain and muscle ARNT-like 1; CG8727 PA; Class E basic helix-loop-helix protein 5; cycle; JAP 3; JAP3; Member of PAS protein 3; Member of PAS superfamily 3; MGC47515; MOP 3; MOP3; PAS domain-containing protein 3; PASD 3; PASD3; TIC;
抗原和靶标
Hair follicles (at protein level). Highly expressed in the adult brain, skeletal muscle and heart.
- O00327 BMAL1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MADQRMDISSTISDFMSPGPTDLLSSSLGTSGVDCNRKRKGSSTDYQESMDTDKDDPHGRLEYTEHQGRIKNAREAHSQIEKRRRDKMNSFIDELASLVPTCNAMSRKLDKLTVLRMAVQHMKTLRGATNPYTEANYKPTFLSDDELKHLILRAADGFLFVVGCDRGKILFVSESVFKILNYSQNDLIGQSLFDYLHPKDIAKVKEQLSSSDTAPRERLIDAKTGLPVKTDITPGPSRLCSGARRSFFCRMKCNRPSVKVEDKDFPSTCSKKKADRKSFCTIHSTGYLKSWPPTKMGLDEDNEPDNEGCNLSCLVAIGRLHSHVVPQPVNGEIRVKSMEYVSRHAIDGKFVFVDQRATAILAYLPQELLGTSCYEYFHQDDIGHLAECHRQVLQTREKITTNCYKFKIKDGSFITLRSRWFSFMNPWTKEVEYIVSTNTVVLANVLEGGDPTFPQLTASPHSMDSMLPSGEGGPKRTHPTVPGIPGGTRAGAGKIGRMIAEEIMEIHRIRGSSPSSCGSSPLNITSTPPPDASSPGGKKILNGGTPDIPSSGLLSGQAQENPGYPYSDSSSILGENPHIGIDMIDNDQGSSSPSNDEAAMAVIMSLLEADAGLGGPVDFSDLPWPL
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. ARNTL/BMAL1 positively regulates myogenesis and negatively regulates adipogenesis via the transcriptional control of the genes of the canonical Wnt signaling pathway. Plays a role in normal pancreatic beta-cell function; regulates glucose-stimulated insulin secretion via the regulation of antioxidant genes NFE2L2/NRF2 and its targets SESN2, PRDX3, CCLC and CCLM. Negatively regulates the mTORC1 signaling pathway; regulates the expression of MTOR and DEPTOR. Controls diurnal oscillations of Ly6C inflammatory monocytes; rhythmic recruitment of the PRC2 complex imparts diurnal variation to chemokine expression that is necessary to sustain Ly6C monocyte rhythms. Regulates the expression of HSD3B2, STAR, PTGS2, CYP11A1, CYP19A1 and LHCGR in the ovary and also the genes involved in hair growth. Plays an important role in adult hippocampal neurogenesis by regulating the timely entry of neural stem/progenitor cells (NSPCs) into the cell cycle and the number of cell divisions that take place prior to cell-cycle exit. Regulates the circadian expression of CIART and KLF11. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The NPAS2-ARNTL/BMAL1 heterodimer positively regulates the expression of MAOA, F7 and LDHA and modulates the circadian rhythm of daytime contrast sensitivity by regulating the rhythmic expression of adenylate cyclase type 1 (ADCY1) in the retina. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence. CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3'. The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3'. Essential for the rhythmic interaction of CLOCK with ASS1 and plays a critical role in positively regulating CLOCK-mediated acetylation of ASS1. Plays a role in protecting against lethal sepsis by limiting the expression of immune checkpoint protein CD274 in macrophages in a PKM2-dependent manner (By similarity). Regulates the diurnal rhythms of skeletal muscle metabolism via transcriptional activation of genes promoting triglyceride synthesis (DGAT2) and metabolic efficiency (COQ10B) (By similarity).
Ubiquitinated, leading to its proteasomal degradation. Deubiquitinated by USP9X.
O-glycosylated; contains O-GlcNAc. O-glycosylation by OGT prevents protein degradation by inhibiting ubiquitination. It also stabilizes the CLOCK-ARNTL/BMAL1 heterodimer thereby increasing CLOCK-ARNTL/BMAL1-mediated transcription of genes in the negative loop of the circadian clock such as PER1/2/3 and CRY1/2.
Acetylated on Lys-538 upon dimerization with CLOCK. Acetylation facilitates CRY1-mediated repression. Deacetylated by SIRT1, which may result in decreased protein stability.
Phosphorylated upon dimerization with CLOCK. Phosphorylation enhances the transcriptional activity, alters the subcellular localization and decreases the stability of the CLOCK-ARNTL/BMAL1 heterodimer by promoting its degradation. Phosphorylation shows circadian variations in the liver with a peak between CT10 to CT14. Phosphorylation at Ser-90 by CK2 is essential for its nuclear localization, its interaction with CLOCK and controls CLOCK nuclear entry (By similarity). Dephosphorylation at Ser-78 is important for dimerization with CLOCK and transcriptional activity.
Sumoylated on Lys-259 upon dimerization with CLOCK. Predominantly conjugated to poly-SUMO2/3 rather than SUMO1 and the level of these conjugates undergo rhythmic variation, peaking at CT9-CT12. Sumoylation localizes it exclusively to the PML body and promotes its ubiquitination in the PML body, ubiquitin-dependent proteasomal degradation and the transcriptional activity of the CLOCK-ARNTL/BMAL1 heterodimer.
Undergoes lysosome-mediated degradation in a time-dependent manner in the liver.
Nucleus. Cytoplasm. Nucleus>PML body.
Note: Shuttles between the nucleus and the cytoplasm and this nucleocytoplasmic shuttling is essential for the nuclear accumulation of CLOCK, target gene transcription and the degradation of the CLOCK-ARNTL/BMAL1 heterodimer. The sumoylated form localizes in the PML body. Sequestered to the cytoplasm in the presence of ID2.
Hair follicles (at protein level). Highly expressed in the adult brain, skeletal muscle and heart.
Component of the circadian clock oscillator which includes the CRY1/2 proteins, CLOCK or NPAS2, ARNTL/BMAL1 or ARNTL2/BMAL2, CSNK1D and/or CSNK1E, TIMELESS and the PER1/2/3 proteins (By similarity). Forms a heterodimer with CLOCK. The CLOCK-ARNTL/BMAL1 heterodimer is required for E-box-dependent transactivation, for CLOCK nuclear translocation and degradation, and, for phosphorylation of both CLOCK and ARNTL/BMAL1 (By similarity). Part of a nuclear complex which also includes RACK1 and PRKCA; RACK1 and PRKCA are recruited to the complex in a circadian manner (By similarity). Interacts with NPAS2 (By similarity). Interacts with EZH2 (By similarity). Interacts with SUMO3 (By similarity). Interacts with SIRT1 (By similarity). Interacts with AHR. Interacts with ID1, ID2 and ID3 (By similarity). Interacts with DDX4 (By similarity). Interacts with OGT (By similarity). Interacts with EED and SUZ12 (By similarity). Interacts with MTA1 (By similarity). Interacts with CIART. Interacts with HSP90. Interacts with KAT2B and EP300. Interacts with BHLHE40/DEC1 and BHLHE41/DEC2 (By similarity). Interacts with RELB and the interaction is enhanced in the presence of CLOCK (By similarity). Interacts with PER1, PER2, CRY1 and CRY2 and this interaction requires a translocation to the nucleus (By similarity). Interaction of the CLOCK-ARNTL/BMAL1 heterodimer with PER or CRY inhibits transcription activation (By similarity). Interaction of the CLOCK-ARNTL/BMAL1 with CRY1 is independent of DNA but with PER2 is off DNA (By similarity). The CLOCK-ARNTL/BMAL1 heterodimer interacts with GSK3B (By similarity). Interacts with KDM5A. Interacts with KMT2A; in a circadian manner (By similarity). Interacts with UBE3A. Interacts with PRKCG (By similarity). Interacts with MAGEL2 (By similarity). Interacts with NCOA2 (By similarity). Interacts with THRAP3 (By similarity). The CLOCK-ARNTL/BMAL1 heterodimer interacts with PASD1. Interacts with PASD1. Interacts with USP9X. Interacts with PIWIL2 (via PIWI domain). Interacts with HDAC3 (By similarity). Interacts with HNF4A.
研究领域
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Organismal Systems > Environmental adaptation > Circadian rhythm. (View pathway)
· Organismal Systems > Nervous system > Dopaminergic synapse.
文献引用
Application: WB Species: Mice Sample: liver tissue and RAW264.7 cells
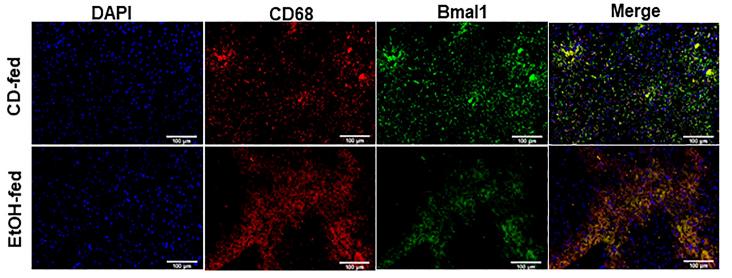
Application: IF/ICC Species: Mice Sample: liver tissue and RAW264.7 cells
Application: IHC Species: Mice Sample: liver tissue and RAW264.7 cells
Application: WB Species: Rat Sample:
Application: WB Species: Mouse Sample:
Application: IF/ICC Species: Mouse Sample:
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.