| 产品: | 磷酸化 Insulin Receptor beta (Tyr1361) 抗体 |
| 货号: | AF3099 |
| 描述: | Rabbit polyclonal antibody to Phospho-Insulin Receptor beta (Tyr1361) |
| 应用: | WB IHC IF/ICC |
| 文献验证: | WB, IF/ICC |
| 反应: | Human, Mouse, Rat |
| 预测: | Pig, Bovine, Sheep, Rabbit, Dog, Chicken, Xenopus |
| 分子量: | 95kDa; 156kD(Calculated). |
| 蛋白号: | P06213 |
| RRID: | AB_2834536 |
产品描述
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# AF3099, RRID:AB_2834536.
展开/折叠
CD220; HHF5; human insulin receptor; Insr; INSR_HUMAN; Insulin receptor subunit beta; IR 1; IR; IR-1; IR1;
抗原和靶标
A synthesized peptide derived from human IR around the phosphorylation site of Tyr1361.
Isoform Long and isoform Short are predominantly expressed in tissue targets of insulin metabolic effects: liver, adipose tissue and skeletal muscle but are also expressed in the peripheral nerve, kidney, pulmonary alveoli, pancreatic acini, placenta vascular endothelium, fibroblasts, monocytes, granulocytes, erythrocytes and skin. Isoform Short is preferentially expressed in fetal cells such as fetal fibroblasts, muscle, liver and kidney. Found as a hybrid receptor with IGF1R in muscle, heart, kidney, adipose tissue, skeletal muscle, hepatoma, fibroblasts, spleen and placenta (at protein level). Overexpressed in several tumors, including breast, colon, lung, ovary, and thyroid carcinomas.
- P06213 INSR_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MATGGRRGAAAAPLLVAVAALLLGAAGHLYPGEVCPGMDIRNNLTRLHELENCSVIEGHLQILLMFKTRPEDFRDLSFPKLIMITDYLLLFRVYGLESLKDLFPNLTVIRGSRLFFNYALVIFEMVHLKELGLYNLMNITRGSVRIEKNNELCYLATIDWSRILDSVEDNYIVLNKDDNEECGDICPGTAKGKTNCPATVINGQFVERCWTHSHCQKVCPTICKSHGCTAEGLCCHSECLGNCSQPDDPTKCVACRNFYLDGRCVETCPPPYYHFQDWRCVNFSFCQDLHHKCKNSRRQGCHQYVIHNNKCIPECPSGYTMNSSNLLCTPCLGPCPKVCHLLEGEKTIDSVTSAQELRGCTVINGSLIINIRGGNNLAAELEANLGLIEEISGYLKIRRSYALVSLSFFRKLRLIRGETLEIGNYSFYALDNQNLRQLWDWSKHNLTITQGKLFFHYNPKLCLSEIHKMEEVSGTKGRQERNDIALKTNGDQASCENELLKFSYIRTSFDKILLRWEPYWPPDFRDLLGFMLFYKEAPYQNVTEFDGQDACGSNSWTVVDIDPPLRSNDPKSQNHPGWLMRGLKPWTQYAIFVKTLVTFSDERRTYGAKSDIIYVQTDATNPSVPLDPISVSNSSSQIILKWKPPSDPNGNITHYLVFWERQAEDSELFELDYCLKGLKLPSRTWSPPFESEDSQKHNQSEYEDSAGECCSCPKTDSQILKELEESSFRKTFEDYLHNVVFVPRKTSSGTGAEDPRPSRKRRSLGDVGNVTVAVPTVAAFPNTSSTSVPTSPEEHRPFEKVVNKESLVISGLRHFTGYRIELQACNQDTPEERCSVAAYVSARTMPEAKADDIVGPVTHEIFENNVVHLMWQEPKEPNGLIVLYEVSYRRYGDEELHLCVSRKHFALERGCRLRGLSPGNYSVRIRATSLAGNGSWTEPTYFYVTDYLDVPSNIAKIIIGPLIFVFLFSVVIGSIYLFLRKRQPDGPLGPLYASSNPEYLSASDVFPCSVYVPDEWEVSREKITLLRELGQGSFGMVYEGNARDIIKGEAETRVAVKTVNESASLRERIEFLNEASVMKGFTCHHVVRLLGVVSKGQPTLVVMELMAHGDLKSYLRSLRPEAENNPGRPPPTLQEMIQMAAEIADGMAYLNAKKFVHRDLAARNCMVAHDFTVKIGDFGMTRDIYETDYYRKGGKGLLPVRWMAPESLKDGVFTTSSDMWSFGVVLWEITSLAEQPYQGLSNEQVLKFVMDGGYLDQPDNCPERVTDLMRMCWQFNPKMRPTFLEIVNLLKDDLHPSFPEVSFFHSEENKAPESEELEMEFEDMENVPLDRSSHCQREEAGGRDGGSSLGFKRSYEEHIPYTHMNGGKKNGRILTLPRSNPS
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Receptor tyrosine kinase which mediates the pleiotropic actions of insulin. Binding of insulin leads to phosphorylation of several intracellular substrates, including, insulin receptor substrates (IRS1, 2, 3, 4), SHC, GAB1, CBL and other signaling intermediates. Each of these phosphorylated proteins serve as docking proteins for other signaling proteins that contain Src-homology-2 domains (SH2 domain) that specifically recognize different phosphotyrosine residues, including the p85 regulatory subunit of PI3K and SHP2. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway, which is responsible for most of the metabolic actions of insulin, and the Ras-MAPK pathway, which regulates expression of some genes and cooperates with the PI3K pathway to control cell growth and differentiation. Binding of the SH2 domains of PI3K to phosphotyrosines on IRS1 leads to the activation of PI3K and the generation of phosphatidylinositol-(3, 4, 5)-triphosphate (PIP3), a lipid second messenger, which activates several PIP3-dependent serine/threonine kinases, such as PDPK1 and subsequently AKT/PKB. The net effect of this pathway is to produce a translocation of the glucose transporter SLC2A4/GLUT4 from cytoplasmic vesicles to the cell membrane to facilitate glucose transport. Moreover, upon insulin stimulation, activated AKT/PKB is responsible for: anti-apoptotic effect of insulin by inducing phosphorylation of BAD; regulates the expression of gluconeogenic and lipogenic enzymes by controlling the activity of the winged helix or forkhead (FOX) class of transcription factors. Another pathway regulated by PI3K-AKT/PKB activation is mTORC1 signaling pathway which regulates cell growth and metabolism and integrates signals from insulin. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 thereby activating mTORC1 pathway. The Ras/RAF/MAP2K/MAPK pathway is mainly involved in mediating cell growth, survival and cellular differentiation of insulin. Phosphorylated IRS1 recruits GRB2/SOS complex, which triggers the activation of the Ras/RAF/MAP2K/MAPK pathway. In addition to binding insulin, the insulin receptor can bind insulin-like growth factors (IGFI and IGFII). Isoform Short has a higher affinity for IGFII binding. When present in a hybrid receptor with IGF1R, binds IGF1.shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by insulin, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2 and insulin. In contrast,shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for insulin. In adipocytes, inhibits lipolysis (By similarity).
After being transported from the endoplasmic reticulum to the Golgi apparatus, the single glycosylated precursor is further glycosylated and then cleaved, followed by its transport to the plasma membrane.
Autophosphorylated on tyrosine residues in response to insulin. Phosphorylation of Tyr-999 is required for binding to IRS1, SHC1 and STAT5B. Dephosphorylated by PTPRE at Tyr-999, Tyr-1185, Tyr-1189 and Tyr-1190. Dephosphorylated by PTPRF and PTPN1. Dephosphorylated by PTPN2; down-regulates insulin-induced signaling.
Cell membrane>Single-pass type I membrane protein. Late endosome. Lysosome.
Note: Binding of insulin to INSR induces internalization and lysosomal degradation of the receptor, a means for downregulating this signaling pathway after stimulation. In the presence of SORL1, internalized INSR molecules are redirected back to the cell surface, thereby preventing their lysosomal catabolism and strengthening insulin signal reception.
Isoform Long and isoform Short are predominantly expressed in tissue targets of insulin metabolic effects: liver, adipose tissue and skeletal muscle but are also expressed in the peripheral nerve, kidney, pulmonary alveoli, pancreatic acini, placenta vascular endothelium, fibroblasts, monocytes, granulocytes, erythrocytes and skin. Isoform Short is preferentially expressed in fetal cells such as fetal fibroblasts, muscle, liver and kidney. Found as a hybrid receptor with IGF1R in muscle, heart, kidney, adipose tissue, skeletal muscle, hepatoma, fibroblasts, spleen and placenta (at protein level). Overexpressed in several tumors, including breast, colon, lung, ovary, and thyroid carcinomas.
The tetrameric insulin receptor binds insulin via non-identical regions from two alpha chains, primarily via the C-terminal region of the first INSR alpha chain. Residues from the leucine-rich N-terminus of the other INSR alpha chain also contribute to this insulin binding site. A secondary insulin-binding site is formed by residues at the junction of fibronectin type-III domain 1 and 2.
Belongs to the protein kinase superfamily. Tyr protein kinase family. Insulin receptor subfamily.
研究领域
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Type II diabetes mellitus.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Ovarian steroidogenesis.
· Organismal Systems > Endocrine system > Regulation of lipolysis in adipocytes.
· Organismal Systems > Excretory system > Aldosterone-regulated sodium reabsorption.
文献引用
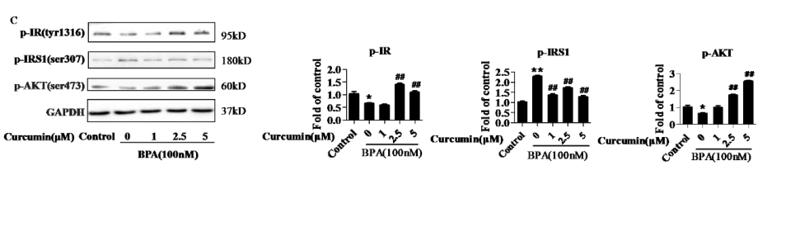
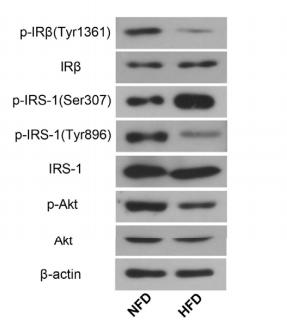
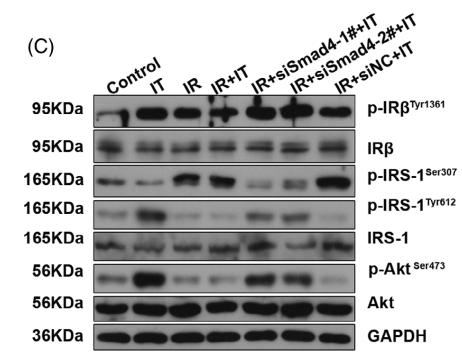
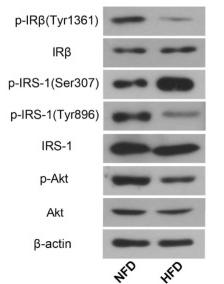
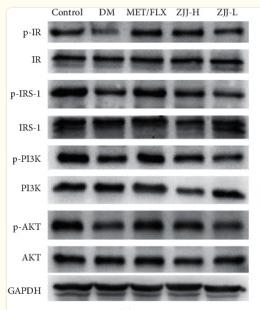
Application: WB Species: Rat Sample:
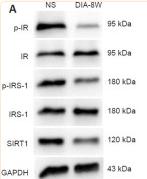
Application: WB Species: Rat Sample: diabetic model rats
Application: WB Species: Human Sample: adipose tissue
Application: WB Species: mouse Sample: placenta
Application: WB Species: Mice Sample: serum and placenta tissues
Application: WB Species: human Sample: HepG2 cells
Application: WB Species: Human Sample: human insulin resistance
Application: WB Species: human Sample: HTR-8/SVneocells
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.