产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# AF3367, RRID:AB_2834782.
展开/折叠
DKFZP586N0721; DKFZp686J10186; hMAD 3; hMAD-3; hSMAD3; HSPC193; HST17436; JV15 2; JV15-2; JV152; LDS1C; LDS3; MAD (mothers against decapentaplegic Drosophila) homolog 3; MAD homolog 3; Mad homolog JV15 2; Mad protein homolog; MAD, mothers against decapentaplegic homolog 3; Mad3; MADH 3; MADH3; MGC60396; Mothers against decapentaplegic homolog 3; Mothers against DPP homolog 3; SMA and MAD related protein 3; SMAD 3; SMAD; SMAD family member 3; SMAD, mothers against DPP homolog 3; Smad3; SMAD3_HUMAN; Drosophila, homolog of, MADR2; hMAD-2; HsMAD2; JV18; JV18-1; JV181; MAD; MAD homolog 2; MAD Related Protein 2; Mad-related protein 2; MADH2; MADR2; MGC22139; MGC34440; Mother against DPP homolog 2; Mothers against decapentaplegic homolog 2; Mothers against decapentaplegic, Drosophila, homolog of, 2; Mothers against DPP homolog 2; OTTHUMP00000163489; Sma and Mad related protein 2; Sma- and Mad-related protein 2 MAD; SMAD 2; SMAD family member 2; SMAD, mothers against DPP homolog 2; SMAD2; SMAD2_HUMAN;
抗原和靶标
A synthesized peptide derived from human Smad2/3 around the phosphorylation site of Thr8.
Expressed at high levels in skeletal muscle, endothelial cells, heart and placenta.
- P84022 SMAD3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSSILPFTPPIVKRLLGWKKGEQNGQEEKWCEKAVKSLVKKLKKTGQLDELEKAITTQNVNTKCITIPRSLDGRLQVSHRKGLPHVIYCRLWRWPDLHSHHELRAMELCEFAFNMKKDEVCVNPYHYQRVETPVLPPVLVPRHTEIPAEFPPLDDYSHSIPENTNFPAGIEPQSNIPETPPPGYLSEDGETSDHQMNHSMDAGSPNLSPNPMSPAHNNLDLQPVTYCEPAFWCSISYYELNQRVGETFHASQPSMTVDGFTDPSNSERFCLGLLSNVNRNAAVELTRRHIGRGVRLYYIGGEVFAECLSDSAIFVQSPNCNQRYGWHPATVCKIPPGCNLKIFNNQEFAALLAQSVNQGFEAVYQLTRMCTIRMSFVKGWGAEYRRQTVTSTPCWIELHLNGPLQWLDKVLTQMGSPSIRCSSVS
- Q15796 SMAD2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSSILPFTPPVVKRLLGWKKSAGGSGGAGGGEQNGQEEKWCEKAVKSLVKKLKKTGRLDELEKAITTQNCNTKCVTIPSTCSEIWGLSTPNTIDQWDTTGLYSFSEQTRSLDGRLQVSHRKGLPHVIYCRLWRWPDLHSHHELKAIENCEYAFNLKKDEVCVNPYHYQRVETPVLPPVLVPRHTEILTELPPLDDYTHSIPENTNFPAGIEPQSNYIPETPPPGYISEDGETSDQQLNQSMDTGSPAELSPTTLSPVNHSLDLQPVTYSEPAFWCSIAYYELNQRVGETFHASQPSLTVDGFTDPSNSERFCLGLLSNVNRNATVEMTRRHIGRGVRLYYIGGEVFAECLSDSAIFVQSPNCNQRYGWHPATVCKIPPGCNLKIFNNQEFAALLAQSVNQGFEAVYQLTRMCTIRMSFVKGWGAEYRRQTVTSTPCWIELHLNGPLQWLDKVLTQMGSPSVRCSSMS
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Receptor-regulated SMAD (R-SMAD) that is an intracellular signal transducer and transcriptional modulator activated by TGF-beta (transforming growth factor) and activin type 1 receptor kinases. Binds the TRE element in the promoter region of many genes that are regulated by TGF-beta and, on formation of the SMAD3/SMAD4 complex, activates transcription. Also can form a SMAD3/SMAD4/JUN/FOS complex at the AP-1/SMAD site to regulate TGF-beta-mediated transcription. Has an inhibitory effect on wound healing probably by modulating both growth and migration of primary keratinocytes and by altering the TGF-mediated chemotaxis of monocytes. This effect on wound healing appears to be hormone-sensitive. Regulator of chondrogenesis and osteogenesis and inhibits early healing of bone fractures. Positively regulates PDPK1 kinase activity by stimulating its dissociation from the 14-3-3 protein YWHAQ which acts as a negative regulator.
Phosphorylated on serine and threonine residues. Enhanced phosphorylation in the linker region on Thr-179, Ser-204 and Ser-208 on EGF and TGF-beta treatment. Ser-208 is the main site of MAPK-mediated phosphorylation. CDK-mediated phosphorylation occurs in a cell-cycle dependent manner and inhibits both the transcriptional activity and antiproliferative functions of SMAD3. This phosphorylation is inhibited by flavopiridol. Maximum phosphorylation at the G(1)/S junction. Also phosphorylated on serine residues in the C-terminal SXS motif by TGFBR1 and ACVR1. TGFBR1-mediated phosphorylation at these C-terminal sites is required for interaction with SMAD4, nuclear location and transactivational activity, and appears to be a prerequisite for the TGF-beta mediated phosphorylation in the linker region. Dephosphorylated in the C-terminal SXS motif by PPM1A. This dephosphorylation disrupts the interaction with SMAD4, promotes nuclear export and terminates TGF-beta-mediated signaling. Phosphorylation at Ser-418 by CSNK1G2/CK1 promotes ligand-dependent ubiquitination and subsequent proteasome degradation, thus inhibiting SMAD3-mediated TGF-beta responses. Phosphorylated by PDPK1.
Acetylation in the nucleus by EP300 in the MH2 domain regulates positively its transcriptional activity and is enhanced by TGF-beta.
Poly-ADP-ribosylated by PARP1 and PARP2. ADP-ribosylation negatively regulates SMAD3 transcriptional responses during the course of TGF-beta signaling.
Ubiquitinated. Monoubiquitinated, leading to prevent DNA-binding. Deubiquitination by USP15 alleviates inhibition and promotes activation of TGF-beta target genes. Ubiquitinated by RNF111, leading to its degradation: only SMAD3 proteins that are 'in use' are targeted by RNF111, RNF111 playing a key role in activating SMAD3 and regulating its turnover (By similarity). Undergoes STUB1-mediated ubiquitination and degradation.
Cytoplasm. Nucleus.
Note: Cytoplasmic and nuclear in the absence of TGF-beta. On TGF-beta stimulation, migrates to the nucleus when complexed with SMAD4 (PubMed:15799969). Through the action of the phosphatase PPM1A, released from the SMAD2/SMAD4 complex, and exported out of the nucleus by interaction with RANBP1 (PubMed:16751101, PubMed:19289081). Co-localizes with LEMD3 at the nucleus inner membrane (PubMed:15601644). MAPK-mediated phosphorylation appears to have no effect on nuclear import (PubMed:19218245). PDPK1 prevents its nuclear translocation in response to TGF-beta (PubMed:17327236).
The MH1 domain is required for DNA binding. Also binds zinc ions which are necessary for the DNA binding.
The MH2 domain is required for both homomeric and heteromeric interactions and for transcriptional regulation. Sufficient for nuclear import.
The linker region is required for the TGFbeta-mediated transcriptional activity and acts synergistically with the MH2 domain.
Belongs to the dwarfin/SMAD family.
Receptor-regulated SMAD (R-SMAD) that is an intracellular signal transducer and transcriptional modulator activated by TGF-beta (transforming growth factor) and activin type 1 receptor kinases. Binds the TRE element in the promoter region of many genes that are regulated by TGF-beta and, on formation of the SMAD2/SMAD4 complex, activates transcription. May act as a tumor suppressor in colorectal carcinoma. Positively regulates PDPK1 kinase activity by stimulating its dissociation from the 14-3-3 protein YWHAQ which acts as a negative regulator.
Phosphorylated on one or several of Thr-220, Ser-245, Ser-250, and Ser-255. In response to TGF-beta, phosphorylated on Ser-465/467 by TGF-beta and activin type 1 receptor kinases. TGF-beta-induced Ser-465/467 phosphorylation declines progressively in a KMT5A-dependent manner. Able to interact with SMURF2 when phosphorylated on Ser-465/467, recruiting other proteins, such as SNON, for degradation. In response to decorin, the naturally occurring inhibitor of TGF-beta signaling, phosphorylated on Ser-240 by CaMK2. Phosphorylated by MAPK3 upon EGF stimulation; which increases transcriptional activity and stability, and is blocked by calmodulin. Phosphorylated by PDPK1.
In response to TGF-beta, ubiquitinated by NEDD4L; which promotes its degradation. Monoubiquitinated, leading to prevent DNA-binding (By similarity). Deubiquitination by USP15 alleviates inhibition and promotes activation of TGF-beta target genes. Ubiquitinated by RNF111, leading to its degradation: only SMAD2 proteins that are 'in use' are targeted by RNF111, RNF111 playing a key role in activating SMAD2 and regulating its turnover (By similarity).
Acetylated on Lys-19 by coactivators in response to TGF-beta signaling, which increases transcriptional activity. Isoform short: Acetylation increases DNA binding activity in vitro and enhances its association with target promoters in vivo. Acetylation in the nucleus by EP300 is enhanced by TGF-beta.
Cytoplasm. Nucleus.
Note: Cytoplasmic and nuclear in the absence of TGF-beta. On TGF-beta stimulation, migrates to the nucleus when complexed with SMAD4 (PubMed:9865696). On dephosphorylation by phosphatase PPM1A, released from the SMAD2/SMAD4 complex, and exported out of the nucleus by interaction with RANBP1 (PubMed:16751101, PubMed:19289081).
Expressed at high levels in skeletal muscle, endothelial cells, heart and placenta.
Belongs to the dwarfin/SMAD family.
研究领域
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Immune diseases > Inflammatory bowel disease (IBD).
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
· Organismal Systems > Endocrine system > Relaxin signaling pathway.
文献引用
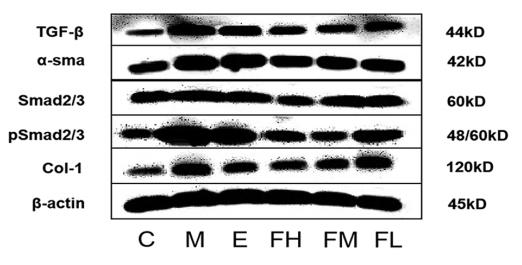
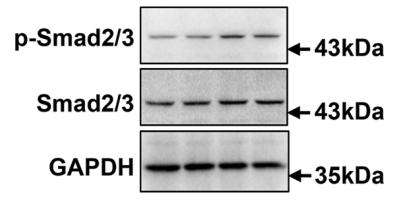
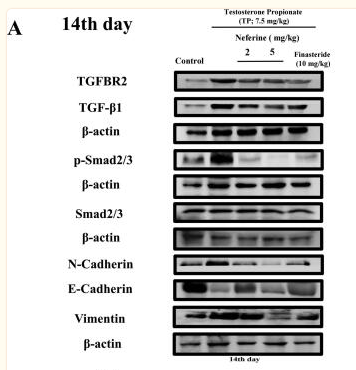
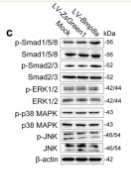
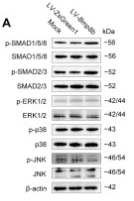
Application: IHC Species: Rat Sample: abdominal aortic tissues
Application: WB Species: Mouse Sample: 3T3-L1 cells
Application: WB Species: Mouse Sample:
Application: WB Species: Mouse Sample: fibroblasts
Application: WB Species: Mouse Sample: liver tissue
Application: IHC Species: Mouse Sample: liver tissue
Application: WB Species: Human Sample:
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.