PCNA Antibody - #AF0239
产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# AF0239, RRID:AB_2833414.
展开/折叠
ATLD2; cb16; Cyclin; DNA polymerase delta auxiliary protein; etID36690.10; fa28e03; fb36g03; HGCN8729; MGC8367; Mutagen-sensitive 209 protein; OTTHUMP00000030189; OTTHUMP00000030190; PCNA; Pcna/cyclin; PCNA_HUMAN; PCNAR; Polymerase delta accessory protein; Proliferating cell nuclear antigen; wu:fa28e03; wu:fb36g03;
抗原和靶标
- P12004 PCNA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MFEARLVQGSILKKVLEALKDLINEACWDISSSGVNLQSMDSSHVSLVQLTLRSEGFDTYRCDRNLAMGVNLTSMSKILKCAGNEDIITLRAEDNADTLALVFEAPNQEKVSDYEMKLMDLDVEQLGIPEQEYSCVVKMPSGEFARICRDLSHIGDAVVISCAKDGVKFSASGELGNGNIKLSQTSNVDKEEEAVTIEMNEPVQLTFALRYLNFFTKATPLSSTVTLSMSADVPLVVEYKIADMGHLKYYLAPKIEDEEGS
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand. Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways. Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion.
Phosphorylated. Phosphorylation at Tyr-211 by EGFR stabilizes chromatin-associated PCNA.
Acetylated by CREBBP and p300/EP300; preferentially acetylated by CREBBP on Lys-80, Lys-13 and Lys-14 and on Lys-77 by p300/EP300 upon loading on chromatin in response to UV irradiation. Lysine acetylation disrupts association with chromatin, hence promoting PCNA ubiquitination and proteasomal degradation in response to UV damage in a CREBBP- and EP300-dependent manner. Acetylation disrupts interaction with NUDT15 and promotes degradation.
Ubiquitinated. Following DNA damage, can be either monoubiquitinated to stimulate direct bypass of DNA lesions by specialized DNA polymerases or polyubiquitinated to promote recombination-dependent DNA synthesis across DNA lesions by template switching mechanisms. Following induction of replication stress, monoubiquitinated by the UBE2B-RAD18 complex on Lys-164, leading to recruit translesion (TLS) polymerases, which are able to synthesize across DNA lesions in a potentially error-prone manner. An error-free pathway also exists and requires non-canonical polyubiquitination on Lys-164 through 'Lys-63' linkage of ubiquitin moieties by the E2 complex UBE2N-UBE2V2 and the E3 ligases, HLTF, RNF8 and SHPRH. This error-free pathway, also known as template switching, employs recombination mechanisms to synthesize across the lesion, using as a template the undamaged, newly synthesized strand of the sister chromatid. Monoubiquitination at Lys-164 also takes place in undamaged proliferating cells, and is mediated by the DCX(DTL) complex, leading to enhance PCNA-dependent translesion DNA synthesis. Sumoylated during S phase.
Methylated on glutamate residues by ARMT1/C6orf211.
Nucleus.
Note: Colocalizes with CREBBP, EP300 and POLD1 to sites of DNA damage (PubMed:24939902). Forms nuclear foci representing sites of ongoing DNA replication and vary in morphology and number during S phase. Together with APEX2, is redistributed in discrete nuclear foci in presence of oxidative DNA damaging agents.
Homotrimer. Interacts with p300/EP300; the interaction occurs on chromatin in UV-irradiated damaged cells. Interacts with CREBBP (via transactivation domain and C-terminus); the interaction occurs on chromatin in UV-irradiated damaged cells. Directly interacts with POLD1, POLD3 and POLD4 subunits of the DNA polymerase delta complex, POLD3 being the major interacting partner; the interaction with POLD3 is inhibited by CDKN1A/p21(CIP1). Forms a complex with activator 1 heteropentamer in the presence of ATP. Interacts with EXO1, POLH, POLK, DNMT1, ERCC5, FEN1, CDC6 and POLDIP2. Interacts with APEX2; this interaction is triggered by reactive oxygen species and increased by misincorporation of uracil in nuclear DNA. Forms a ternary complex with DNTTIP2 and core histone. Interacts with KCTD10 and PPP1R15A (By similarity). Directly interacts with BAZ1B. Interacts with HLTF and SHPRH. Interacts with NUDT15; this interaction is disrupted in response to UV irradiation and acetylation. Interacts with CDKN1A/p21(CIP1) and CDT1; interacts via their PIP-box which also recruits the DCX(DTL) complex. The interaction with CDKN1A inhibits POLD3 binding. Interacts with DDX11. Interacts with EGFR; positively regulates PCNA. Interacts with PARPBP. Interacts (when ubiquitinated) with SPRTN; leading to enhance RAD18-mediated PCNA ubiquitination. Interacts (when polyubiquitinated) with ZRANB3. Interacts with SMARCAD1. Interacts with CDKN1C. Interacts with PCLAF (via PIP-box). Interacts with RTEL1 (via PIP-box); the interaction is direct and essential for the suppression of telomere fragility. Interacts with FAM111A (via PIP-box); the interaction is direct and required for PCNA loading on chromatin binding. Interacts with LIG1. Interacts with SETMAR. Interacts with ANKRD17. Interacts with FBXO18/FBH1 (via PIP-box); the interaction recruits the DCX(DTL) complex and promotes ubiquitination and degradation of FBXO18/FBH1. Interacts with POLN. Interacts with SDE2 (via PIP-box); the interaction is direct and prevents ultraviolet light induced monoubiquitination. Component of the replisome complex composed of at least DONSON, MCM2, MCM7, PCNA and TICRR; interaction at least with PCNA occurs during DNA replication. Interacts with MAPK15; the interaction is chromatin binding dependent and prevents MDM2-mediated PCNA destruction by inhibiting the association of PCNA with MDM2. Interacts with PARP10 (via PIP-box). Interacts with DDI2. Interacts with HMCES (via PIP-box).
(Microbial infection) Interacts with herpes virus 8 protein LANA1.
Belongs to the PCNA family.
研究领域
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Genetic Information Processing > Replication and repair > DNA replication.
· Genetic Information Processing > Replication and repair > Base excision repair.
· Genetic Information Processing > Replication and repair > Nucleotide excision repair.
· Genetic Information Processing > Replication and repair > Mismatch repair.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
文献引用
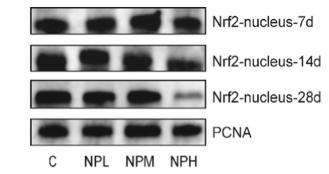
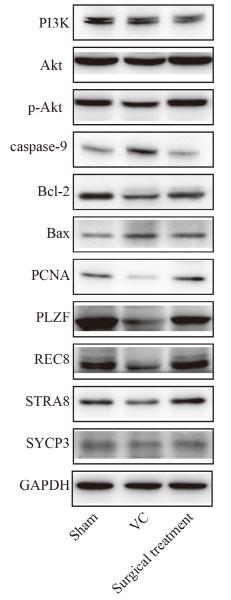
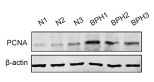
Application: WB Species: Rat Sample:
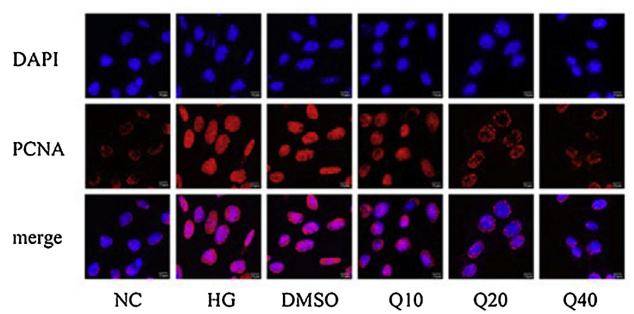
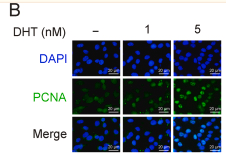
Application: IF/ICC Species: Rat Sample:
Application: IF/ICC Species: mouse Sample: glomerular mesangial cells
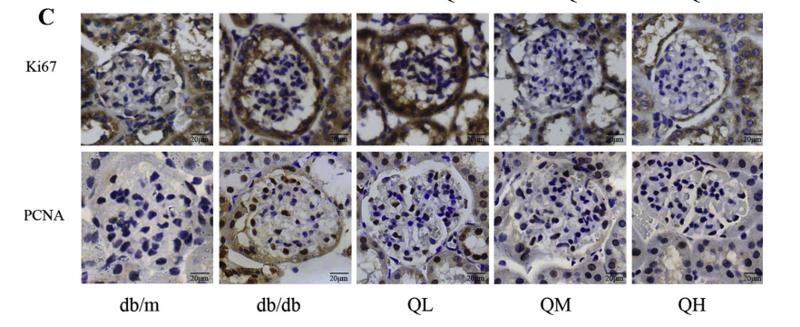
Application: IHC Species: mouse Sample: renal cortex
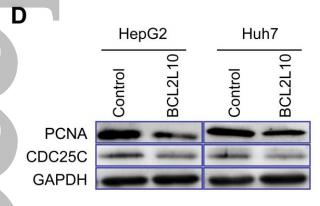
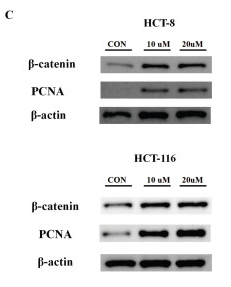
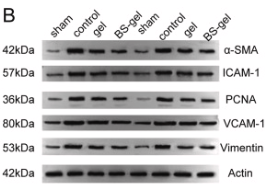
Application: WB Species: Human Sample: HCT 8 cells and HCT 116 cells
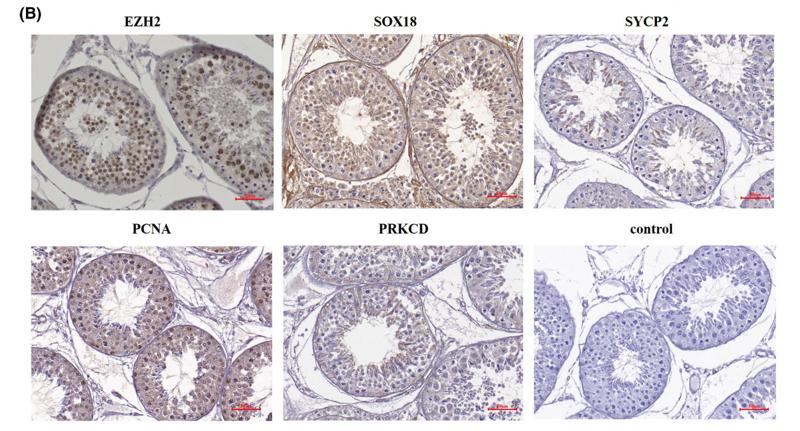
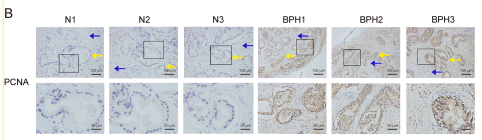
Application: IHC Species: Rat Sample: liver tissue
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.