| 产品: | pan Sodium Channel 抗体 |
| 货号: | AF0255 |
| 描述: | Rabbit polyclonal antibody to pan Sodium Channel |
| 应用: | WB IHC IF/ICC |
| 文献验证: | WB |
| 反应: | Human, Mouse, Rat |
| 预测: | Bovine, Horse, Sheep, Rabbit, Chicken |
| 分子量: | 230kDa; 229kD,208kD,227kD,226kD,228kD,205kD,225kD,221kD(Calculated). |
| 蛋白号: | P35498 | P35499 | Q14524 | Q15858 | Q99250 | Q9NY46 | Q9UI33 | Q9UQD0 | Q9Y5Y9 |
| RRID: | AB_2833430 |
产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# AF0255, RRID:AB_2833430.
展开/折叠
HBSC II;HH1;hNaN;hNE-Na;hPN3;KIAA1356;MED;NAC1;NAC2;NAC3;NENA;Neuroendocrine sodium channel;Peripheral nerve sodium channel 3;Peripheral nerve sodium channel 5;Peripheral sodium channel 1;PN1;PN3;PN5;SCN1;SCN10A;SCN11A;SCN12A;SCN1A;SCN2A;SCN2A1;SCN2A2;SCN3A;SCN4A;SCN5A;SCN8A;SCN9A;Sensory neuron sodium channel 2;SkM1;SNS2;Sodium channel protein brain I subunit alpha;Sodium channel protein brain II subunit alpha;Sodium channel protein brain III subunit alpha;Sodium channel protein cardiac muscle subunit alpha;Sodium channel protein skeletal muscle subunit alpha;Sodium channel protein type 1 subunit alpha;Sodium channel protein type 10 subunit alpha;Sodium channel protein type 11 subunit alpha;Sodium channel protein type 2 subunit alpha;Sodium channel protein type 3 subunit alpha;Sodium channel protein type 4 subunit alpha;Sodium channel protein type 5 subunit alpha;Sodium channel protein type 8 subunit alpha;Sodium channel protein type 9 subunit alpha;Sodium channel protein type II subunit alpha;Sodium channel protein type III subunit alpha;Sodium channel protein type IV subunit alpha;Sodium channel protein type IX subunit alpha;Sodium channel protein type V subunit alpha;Sodium channel protein type VIII subunit alpha;Sodium channel protein type X subunit alpha;Sodium channel protein type XI subunit alpha;Voltage-gated sodium channel subtype III;Voltage-gated sodium channel subunit alpha Nav1.1;Voltage-gated sodium channel subunit alpha Nav1.2;Voltage-gated sodium channel subunit alpha Nav1.3;Voltage-gated sodium channel subunit alpha Nav1.4;Voltage-gated sodium channel subunit alpha Nav1.5;Voltage-gated sodium channel subunit alpha Nav1.6;Voltage-gated sodium channel subunit alpha Nav1.7;Voltage-gated sodium channel subunit alpha Nav1.8;Voltage-gated sodium channel subunit alpha Nav1.9;
抗原和靶标
P35498(SCN1A_HUMAN) >>Visit HPA database.
P35499(SCN4A_HUMAN) >>Visit HPA database.
Q14524(SCN5A_HUMAN) >>Visit HPA database.
Q15858(SCN9A_HUMAN) >>Visit HPA database.
Q99250(SCN2A_HUMAN) >>Visit HPA database.
Q9NY46(SCN3A_HUMAN) >>Visit HPA database.
Q9UI33(SCNBA_HUMAN) >>Visit HPA database.
Found in jejunal circular smooth muscle cells (at protein level). Expressed in human atrial and ventricular cardiac muscle but not in adult skeletal muscle, brain, myometrium, liver, or spleen. Isoform 4 is expressed in brain.
Q15858 SCN9A_HUMAN:Expressed strongly in dorsal root ganglion, with only minor levels elsewhere in the body, smooth muscle cells, MTC cell line and C-cell carcinoma. Isoform 1 is expressed preferentially in the central and peripheral nervous system. Isoform 2 is expressed preferentially in the dorsal root ganglion.
Q9NY46 SCN3A_HUMAN:Expressed in enterochromaffin cells in both colon and small bowel (at protein level).
Q9UI33 SCNBA_HUMAN:Expressed in the dorsal root ganglia and trigeminal ganglia, olfactory bulb, hippocampus, cerebellar cortex, spinal cord, spleen, small intestine and placenta.
Q9UQD0 SCN8A_HUMAN:Expressed in the hippocampus with increased expression in epileptic tissue compared to normal adjacent tissue (at protein level) (PubMed:28842554). Isoform 5: Expressed in non-neuronal tissues, such as monocytes/macrophages.
Q9Y5Y9 SCNAA_HUMAN:Expressed in the dorsal root ganglia and sciatic nerve.
- P35498 SCN1A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEQTVLVPPGPDSFNFFTRESLAAIERRIAEEKAKNPKPDKKDDDENGPKPNSDLEAGKNLPFIYGDIPPEMVSEPLEDLDPYYINKKTFIVLNKGKAIFRFSATSALYILTPFNPLRKIAIKILVHSLFSMLIMCTILTNCVFMTMSNPPDWTKNVEYTFTGIYTFESLIKIIARGFCLEDFTFLRDPWNWLDFTVITFAYVTEFVDLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCIQWPPTNASLEEHSIEKNITVNYNGTLINETVFEFDWKSYIQDSRYHYFLEGFLDALLCGNSSDAGQCPEGYMCVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDFWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLINLILAVVAMAYEEQNQATLEEAEQKEAEFQQMIEQLKKQQEAAQQAATATASEHSREPSAAGRLSDSSSEASKLSSKSAKERRNRRKKRKQKEQSGGEEKDEDEFQKSESEDSIRRKGFRFSIEGNRLTYEKRYSSPHQSLLSIRGSLFSPRRNSRTSLFSFRGRAKDVGSENDFADDEHSTFEDNESRRDSLFVPRRHGERRNSNLSQTSRSSRMLAVFPANGKMHSTVDCNGVVSLVGGPSVPTSPVGQLLPEVIIDKPATDDNGTTTETEMRKRRSSSFHVSMDFLEDPSQRQRAMSIASILTNTVEELEESRQKCPPCWYKFSNIFLIWDCSPYWLKVKHVVNLVVMDPFVDLAITICIVLNTLFMAMEHYPMTDHFNNVLTVGNLVFTGIFTAEMFLKIIAMDPYYYFQEGWNIFDGFIVTLSLVELGLANVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKDCVCKIASDCQLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLTVFMMVMVIGNLVVLNLFLALLLSSFSADNLAATDDDNEMNNLQIAVDRMHKGVAYVKRKIYEFIQQSFIRKQKILDEIKPLDDLNNKKDSCMSNHTAEIGKDLDYLKDVNGTTSGIGTGSSVEKYIIDESDYMSFINNPSLTVTVPIAVGESDFENLNTEDFSSESDLEESKEKLNESSSSSEGSTVDIGAPVEEQPVVEPEETLEPEACFTEGCVQRFKCCQINVEEGRGKQWWNLRRTCFRIVEHNWFETFIVFMILLSSGALAFEDIYIDQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGYQTYFTNAWCWLDFLIVDVSLVSLTANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCINTTTGDRFDIEDVNNHTDCLKLIERNETARWKNVKVNFDNVGFGYLSLLQVATFKGWMDIMYAAVDSRNVELQPKYEESLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPGNKFQGMVFDFVTRQVFDISIMILICLNMVTMMVETDDQSEYVTTILSRINLVFIVLFTGECVLKLISLRHYYFTIGWNIFDFVVVILSIVGMFLAELIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKREVGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSKPPDCDPNKVNPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFMEFEKLSQFAAALEPPLNLPQPNKLQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEERFMASNPSKVSYQPITTTLKRKQEEVSAVIIQRAYRRHLLKRTVKQASFTYNKNKIKGGANLLIKEDMIIDRINENSITEKTDLTMSTAACPPSYDRVTKPIVEKHEQEGKDEKAKGK
- P35499 SCN4A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARPSLCTLVPLGPECLRPFTRESLAAIEQRAVEEEARLQRNKQMEIEEPERKPRSDLEAGKNLPMIYGDPPPEVIGIPLEDLDPYYSNKKTFIVLNKGKAIFRFSATPALYLLSPFSVVRRGAIKVLIHALFSMFIMITILTNCVFMTMSDPPPWSKNVEYTFTGIYTFESLIKILARGFCVDDFTFLRDPWNWLDFSVIMMAYLTEFVDLGNISALRTFRVLRALKTITVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALVGLQLFMGNLRQKCVRWPPPFNDTNTTWYSNDTWYGNDTWYGNEMWYGNDSWYANDTWNSHASWATNDTFDWDAYISDEGNFYFLEGSNDALLCGNSSDAGHCPEGYECIKTGRNPNYGYTSYDTFSWAFLALFRLMTQDYWENLFQLTLRAAGKTYMIFFVVIIFLGSFYLINLILAVVAMAYAEQNEATLAEDKEKEEEFQQMLEKFKKHQEELEKAKAAQALEGGEADGDPAHGKDCNGSLDTSQGEKGAPRQSSSGDSGISDAMEELEEAHQKCPPWWYKCAHKVLIWNCCAPWLKFKNIIHLIVMDPFVDLGITICIVLNTLFMAMEHYPMTEHFDNVLTVGNLVFTGIFTAEMVLKLIAMDPYEYFQQGWNIFDSIIVTLSLVELGLANVQGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKIALDCNLPRWHMHDFFHSFLIVFRILCGEWIETMWDCMEVAGQAMCLTVFLMVMVIGNLVVLNLFLALLLSSFSADSLAASDEDGEMNNLQIAIGRIKLGIGFAKAFLLGLLHGKILSPKDIMLSLGEADGAGEAGEAGETAPEDEKKEPPEEDLKKDNHILNHMGLADGPPSSLELDHLNFINNPYLTIQVPIASEESDLEMPTEEETDTFSEPEDSKKPPQPLYDGNSSVCSTADYKPPEEDPEEQAEENPEGEQPEECFTEACVQRWPCLYVDISQGRGKKWWTLRRACFKIVEHNWFETFIVFMILLSSGALAFEDIYIEQRRVIRTILEYADKVFTYIFIMEMLLKWVAYGFKVYFTNAWCWLDFLIVDVSIISLVANWLGYSELGPIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYYCINTTTSERFDISEVNNKSECESLMHTGQVRWLNVKVNYDNVGLGYLSLLQVATFKGWMDIMYAAVDSREKEEQPQYEVNLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGKDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPQNKIQGMVYDLVTKQAFDITIMILICLNMVTMMVETDNQSQLKVDILYNINMIFIIIFTGECVLKMLALRQYYFTVGWNIFDFVVVILSIVGLALSDLIQKYFVSPTLFRVIRLARIGRVLRLIRGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMSNFAYVKKESGIDDMFNFETFGNSIICLFEITTSAGWDGLLNPILNSGPPDCDPNLENPGTSVKGDCGNPSIGICFFCSYIIISFLIVVNMYIAIILENFNVATEESSEPLGEDDFEMFYETWEKFDPDATQFIAYSRLSDFVDTLQEPLRIAKPNKIKLITLDLPMVPGDKIHCLDILFALTKEVLGDSGEMDALKQTMEEKFMAANPSKVSYEPITTTLKRKHEEVCAIKIQRAYRRHLLQRSMKQASYMYRHSHDGSGDDAPEKEGLLANTMSKMYGHENGNSSSPSPEEKGEAGDAGPTMGLMPISPSDTAWPPAPPPGQTVRPGVKESLV
- Q14524 SCN5A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MANFLLPRGTSSFRRFTRESLAAIEKRMAEKQARGSTTLQESREGLPEEEAPRPQLDLQASKKLPDLYGNPPQELIGEPLEDLDPFYSTQKTFIVLNKGKTIFRFSATNALYVLSPFHPIRRAAVKILVHSLFNMLIMCTILTNCVFMAQHDPPPWTKYVEYTFTAIYTFESLVKILARGFCLHAFTFLRDPWNWLDFSVIIMAYTTEFVDLGNVSALRTFRVLRALKTISVISGLKTIVGALIQSVKKLADVMVLTVFCLSVFALIGLQLFMGNLRHKCVRNFTALNGTNGSVEADGLVWESLDLYLSDPENYLLKNGTSDVLLCGNSSDAGTCPEGYRCLKAGENPDHGYTSFDSFAWAFLALFRLMTQDCWERLYQQTLRSAGKIYMIFFMLVIFLGSFYLVNLILAVVAMAYEEQNQATIAETEEKEKRFQEAMEMLKKEHEALTIRGVDTVSRSSLEMSPLAPVNSHERRSKRRKRMSSGTEECGEDRLPKSDSEDGPRAMNHLSLTRGLSRTSMKPRSSRGSIFTFRRRDLGSEADFADDENSTAGESESHHTSLLVPWPLRRTSAQGQPSPGTSAPGHALHGKKNSTVDCNGVVSLLGAGDPEATSPGSHLLRPVMLEHPPDTTTPSEEPGGPQMLTSQAPCVDGFEEPGARQRALSAVSVLTSALEELEESRHKCPPCWNRLAQRYLIWECCPLWMSIKQGVKLVVMDPFTDLTITMCIVLNTLFMALEHYNMTSEFEEMLQVGNLVFTGIFTAEMTFKIIALDPYYYFQQGWNIFDSIIVILSLMELGLSRMSNLSVLRSFRLLRVFKLAKSWPTLNTLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKNYSELRDSDSGLLPRWHMMDFFHAFLIIFRILCGEWIETMWDCMEVSGQSLCLLVFLLVMVIGNLVVLNLFLALLLSSFSADNLTAPDEDREMNNLQLALARIQRGLRFVKRTTWDFCCGLLRQRPQKPAALAAQGQLPSCIATPYSPPPPETEKVPPTRKETRFEEGEQPGQGTPGDPEPVCVPIAVAESDTDDQEEDEENSLGTEEESSKQQESQPVSGGPEAPPDSRTWSQVSATASSEAEASASQADWRQQWKAEPQAPGCGETPEDSCSEGSTADMTNTAELLEQIPDLGQDVKDPEDCFTEGCVRRCPCCAVDTTQAPGKVWWRLRKTCYHIVEHSWFETFIIFMILLSSGALAFEDIYLEERKTIKVLLEYADKMFTYVFVLEMLLKWVAYGFKKYFTNAWCWLDFLIVDVSLVSLVANTLGFAEMGPIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFGRCINQTEGDLPLNYTIVNNKSQCESLNLTGELYWTKVKVNFDNVGAGYLALLQVATFKGWMDIMYAAVDSRGYEEQPQWEYNLYMYIYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKYQGFIFDIVTKQAFDVTIMFLICLNMVTMMVETDDQSPEKINILAKINLLFVAIFTGECIVKLAALRHYYFTNSWNIFDFVVVILSIVGTVLSDIIQKYFFSPTLFRVIRLARIGRILRLIRGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMANFAYVKWEAGIDDMFNFQTFANSMLCLFQITTSAGWDGLLSPILNTGPPYCDPTLPNSNGSRGDCGSPAVGILFFTTYIIISFLIVVNMYIAIILENFSVATEESTEPLSEDDFDMFYEIWEKFDPEATQFIEYSVLSDFADALSEPLRIAKPNQISLINMDLPMVSGDRIHCMDILFAFTKRVLGESGEMDALKIQMEEKFMAANPSKISYEPITTTLRRKHEEVSAMVIQRAFRRHLLQRSLKHASFLFRQQAGSGLSEEDAPEREGLIAYVMSENFSRPLGPPSSSSISSTSFPPSYDSVTRATSDNLQVRGSDYSHSEDLADFPPSPDRDRESIV
- Q15858 SCN9A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAMLPPPGPQSFVHFTKQSLALIEQRIAERKSKEPKEEKKDDDEEAPKPSSDLEAGKQLPFIYGDIPPGMVSEPLEDLDPYYADKKTFIVLNKGKTIFRFNATPALYMLSPFSPLRRISIKILVHSLFSMLIMCTILTNCIFMTMNNPPDWTKNVEYTFTGIYTFESLVKILARGFCVGEFTFLRDPWNWLDFVVIVFAYLTEFVNLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLKHKCFRNSLENNETLESIMNTLESEEDFRKYFYYLEGSKDALLCGFSTDSGQCPEGYTCVKIGRNPDYGYTSFDTFSWAFLALFRLMTQDYWENLYQQTLRAAGKTYMIFFVVVIFLGSFYLINLILAVVAMAYEEQNQANIEEAKQKELEFQQMLDRLKKEQEEAEAIAAAAAEYTSIRRSRIMGLSESSSETSKLSSKSAKERRNRRKKKNQKKLSSGEEKGDAEKLSKSESEDSIRRKSFHLGVEGHRRAHEKRLSTPNQSPLSIRGSLFSARRSSRTSLFSFKGRGRDIGSETEFADDEHSIFGDNESRRGSLFVPHRPQERRSSNISQASRSPPMLPVNGKMHSAVDCNGVVSLVDGRSALMLPNGQLLPEVIIDKATSDDSGTTNQIHKKRRCSSYLLSEDMLNDPNLRQRAMSRASILTNTVEELEESRQKCPPWWYRFAHKFLIWNCSPYWIKFKKCIYFIVMDPFVDLAITICIVLNTLFMAMEHHPMTEEFKNVLAIGNLVFTGIFAAEMVLKLIAMDPYEYFQVGWNIFDSLIVTLSLVELFLADVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINDDCTLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLIVYMMVMVIGNLVVLNLFLALLLSSFSSDNLTAIEEDPDANNLQIAVTRIKKGINYVKQTLREFILKAFSKKPKISREIRQAEDLNTKKENYISNHTLAEMSKGHNFLKEKDKISGFGSSVDKHLMEDSDGQSFIHNPSLTVTVPIAPGESDLENMNAEELSSDSDSEYSKVRLNRSSSSECSTVDNPLPGEGEEAEAEPMNSDEPEACFTDGCVWRFSCCQVNIESGKGKIWWNIRKTCYKIVEHSWFESFIVLMILLSSGALAFEDIYIERKKTIKIILEYADKIFTYIFILEMLLKWIAYGYKTYFTNAWCWLDFLIVDVSLVTLVANTLGYSDLGPIKSLRTLRALRPLRALSRFEGMRVVVNALIGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYECINTTDGSRFPASQVPNRSECFALMNVSQNVRWKNLKVNFDNVGLGYLSLLQVATFKGWTIIMYAAVDSVNVDKQPKYEYSLYMYIYFVVFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPGNKIQGCIFDLVTNQAFDISIMVLICLNMVTMMVEKEGQSQHMTEVLYWINVVFIILFTGECVLKLISLRHYYFTVGWNIFDFVVVIISIVGMFLADLIETYFVSPTLFRVIRLARIGRILRLVKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKKEDGINDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSKPPDCDPKKVHPGSSVEGDCGNPSVGIFYFVSYIIISFLVVVNMYIAVILENFSVATEESTEPLSEDDFEMFYEVWEKFDPDATQFIEFSKLSDFAAALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDSLRSQMEERFMSANPSKVSYEPITTTLKRKQEDVSATVIQRAYRRYRLRQNVKNISSIYIKDGDRDDDLLNKKDMAFDNVNENSSPEKTDATSSTTSPPSYDSVTKPDKEKYEQDRTEKEDKGKDSKESKK
- Q99250 SCN2A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQSVLVPPGPDSFRFFTRESLAAIEQRIAEEKAKRPKQERKDEDDENGPKPNSDLEAGKSLPFIYGDIPPEMVSVPLEDLDPYYINKKTFIVLNKGKAISRFSATPALYILTPFNPIRKLAIKILVHSLFNMLIMCTILTNCVFMTMSNPPDWTKNVEYTFTGIYTFESLIKILARGFCLEDFTFLRDPWNWLDFTVITFAYVTEFVDLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCLQWPPDNSSFEINITSFFNNSLDGNGTTFNRTVSIFNWDEYIEDKSHFYFLEGQNDALLCGNSSDAGQCPEGYICVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDFWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLINLILAVVAMAYEEQNQATLEEAEQKEAEFQQMLEQLKKQQEEAQAAAAAASAESRDFSGAGGIGVFSESSSVASKLSSKSEKELKNRRKKKKQKEQSGEEEKNDRVRKSESEDSIRRKGFRFSLEGSRLTYEKRFSSPHQSLLSIRGSLFSPRRNSRASLFSFRGRAKDIGSENDFADDEHSTFEDNDSRRDSLFVPHRHGERRHSNVSQASRASRVLPILPMNGKMHSAVDCNGVVSLVGGPSTLTSAGQLLPEGTTTETEIRKRRSSSYHVSMDLLEDPTSRQRAMSIASILTNTMEELEESRQKCPPCWYKFANMCLIWDCCKPWLKVKHLVNLVVMDPFVDLAITICIVLNTLFMAMEHYPMTEQFSSVLSVGNLVFTGIFTAEMFLKIIAMDPYYYFQEGWNIFDGFIVSLSLMELGLANVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKISNDCELPRWHMHDFFHSFLIVFRVLCGEWIETMWDCMEVAGQTMCLTVFMMVMVIGNLVVLNLFLALLLSSFSSDNLAATDDDNEMNNLQIAVGRMQKGIDFVKRKIREFIQKAFVRKQKALDEIKPLEDLNNKKDSCISNHTTIEIGKDLNYLKDGNGTTSGIGSSVEKYVVDESDYMSFINNPSLTVTVPIAVGESDFENLNTEEFSSESDMEESKEKLNATSSSEGSTVDIGAPAEGEQPEVEPEESLEPEACFTEDCVRKFKCCQISIEEGKGKLWWNLRKTCYKIVEHNWFETFIVFMILLSSGALAFEDIYIEQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGFQVYFTNAWCWLDFLIVDVSLVSLTANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCINYTTGEMFDVSVVNNYSECKALIESNQTARWKNVKVNFDNVGLGYLSLLQVATFKGWMDIMYAAVDSRNVELQPKYEDNLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPANKFQGMVFDFVTKQVFDISIMILICLNMVTMMVETDDQSQEMTNILYWINLVFIVLFTGECVLKLISLRYYYFTIGWNIFDFVVVILSIVGMFLAELIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKREVGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSGPPDCDPDKDHPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFIEFAKLSDFADALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEERFMASNPSKVSYEPITTTLKRKQEEVSAIIIQRAYRRYLLKQKVKKVSSIYKKDKGKECDGTPIKEDTLIDKLNENSTPEKTDMTPSTTSPPSYDSVTKPEKEKFEKDKSEKEDKGKDIRESKK
- Q9NY46 SCN3A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQALLVPPGPESFRLFTRESLAAIEKRAAEEKAKKPKKEQDNDDENKPKPNSDLEAGKNLPFIYGDIPPEMVSEPLEDLDPYYINKKTFIVMNKGKAIFRFSATSALYILTPLNPVRKIAIKILVHSLFSMLIMCTILTNCVFMTLSNPPDWTKNVEYTFTGIYTFESLIKILARGFCLEDFTFLRDPWNWLDFSVIVMAYVTEFVSLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCLQWPPSDSAFETNTTSYFNGTMDSNGTFVNVTMSTFNWKDYIGDDSHFYVLDGQKDPLLCGNGSDAGQCPEGYICVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDYWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLVNLILAVVAMAYEEQNQATLEEAEQKEAEFQQMLEQLKKQQEEAQAVAAASAASRDFSGIGGLGELLESSSEASKLSSKSAKEWRNRRKKRRQREHLEGNNKGERDSFPKSESEDSVKRSSFLFSMDGNRLTSDKKFCSPHQSLLSIRGSLFSPRRNSKTSIFSFRGRAKDVGSENDFADDEHSTFEDSESRRDSLFVPHRHGERRNSNVSQASMSSRMVPGLPANGKMHSTVDCNGVVSLVGGPSALTSPTGQLPPEGTTTETEVRKRRLSSYQISMEMLEDSSGRQRAVSIASILTNTMEELEESRQKCPPCWYRFANVFLIWDCCDAWLKVKHLVNLIVMDPFVDLAITICIVLNTLFMAMEHYPMTEQFSSVLTVGNLVFTGIFTAEMVLKIIAMDPYYYFQEGWNIFDGIIVSLSLMELGLSNVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINDDCTLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQTMCLIVFMLVMVIGNLVVLNLFLALLLSSFSSDNLAATDDDNEMNNLQIAVGRMQKGIDYVKNKMRECFQKAFFRKPKVIEIHEGNKIDSCMSNNTGIEISKELNYLRDGNGTTSGVGTGSSVEKYVIDENDYMSFINNPSLTVTVPIAVGESDFENLNTEEFSSESELEESKEKLNATSSSEGSTVDVVLPREGEQAETEPEEDLKPEACFTEGCIKKFPFCQVSTEEGKGKIWWNLRKTCYSIVEHNWFETFIVFMILLSSGALAFEDIYIEQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGFQTYFTNAWCWLDFLIVDVSLVSLVANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCVNMTTGNMFDISDVNNLSDCQALGKQARWKNVKVNFDNVGAGYLALLQVATFKGWMDIMYAAVDSRDVKLQPVYEENLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPANKFQGMVFDFVTRQVFDISIMILICLNMVTMMVETDDQGKYMTLVLSRINLVFIVLFTGEFVLKLVSLRHYYFTIGWNIFDFVVVILSIVGMFLAEMIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKKEAGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSAPPDCDPDTIHPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFIEFSKLSDFAAALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEDRFMASNPSKVSYEPITTTLKRKQEEVSAAIIQRNFRCYLLKQRLKNISSNYNKEAIKGRIDLPIKQDMIIDKLNGNSTPEKTDGSSSTTSPPSYDSVTKPDKEKFEKDKPEKESKGKEVRENQK
- Q9UI33 SCNBA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDDRCYPVIFPDERNFRPFTSDSLAAIEKRIAIQKEKKKSKDQTGEVPQPRPQLDLKASRKLPKLYGDIPRELIGKPLEDLDPFYRNHKTFMVLNRKRTIYRFSAKHALFIFGPFNSIRSLAIRVSVHSLFSMFIIGTVIINCVFMATGPAKNSNSNNTDIAECVFTGIYIFEALIKILARGFILDEFSFLRDPWNWLDSIVIGIAIVSYIPGITIKLLPLRTFRVFRALKAISVVSRLKVIVGALLRSVKKLVNVIILTFFCLSIFALVGQQLFMGSLNLKCISRDCKNISNPEAYDHCFEKKENSPEFKMCGIWMGNSACSIQYECKHTKINPDYNYTNFDNFGWSFLAMFRLMTQDSWEKLYQQTLRTTGLYSVFFFIVVIFLGSFYLINLTLAVVTMAYEEQNKNVAAEIEAKEKMFQEAQQLLKEEKEALVAMGIDRSSLTSLETSYFTPKKRKLFGNKKRKSFFLRESGKDQPPGSDSDEDCQKKPQLLEQTKRLSQNLSLDHFDEHGDPLQRQRALSAVSILTITMKEQEKSQEPCLPCGENLASKYLVWNCCPQWLCVKKVLRTVMTDPFTELAITICIIINTVFLAMEHHKMEASFEKMLNIGNLVFTSIFIAEMCLKIIALDPYHYFRRGWNIFDSIVALLSFADVMNCVLQKRSWPFLRSFRVLRVFKLAKSWPTLNTLIKIIGNSVGALGSLTVVLVIVIFIFSVVGMQLFGRSFNSQKSPKLCNPTGPTVSCLRHWHMGDFWHSFLVVFRILCGEWIENMWECMQEANASSSLCVIVFILITVIGKLVVLNLFIALLLNSFSNEERNGNLEGEARKTKVQLALDRFRRAFCFVRHTLEHFCHKWCRKQNLPQQKEVAGGCAAQSKDIIPLVMEMKRGSETQEELGILTSVPKTLGVRHDWTWLAPLAEEEDDVEFSGEDNAQRITQPEPEQQAYELHQENKKPTSQRVQSVEIDMFSEDEPHLTIQDPRKKSDVTSILSECSTIDLQDGFGWLPEMVPKKQPERCLPKGFGCCFPCCSVDKRKPPWVIWWNLRKTCYQIVKHSWFESFIIFVILLSSGALIFEDVHLENQPKIQELLNCTDIIFTHIFILEMVLKWVAFGFGKYFTSAWCCLDFIIVIVSVTTLINLMELKSFRTLRALRPLRALSQFEGMKVVVNALIGAIPAILNVLLVCLIFWLVFCILGVYFFSGKFGKCINGTDSVINYTIITNKSQCESGNFSWINQKVNFDNVGNAYLALLQVATFKGWMDIIYAAVDSTEKEQQPEFESNSLGYIYFVVFIIFGSFFTLNLFIGVIIDNFNQQQKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKCQGLVFDIVTSQIFDIIIISLIILNMISMMAESYNQPKAMKSILDHLNWVFVVIFTLECLIKIFALRQYYFTNGWNLFDCVVVLLSIVSTMISTLENQEHIPFPPTLFRIVRLARIGRILRLVRAARGIRTLLFALMMSLPSLFNIGLLLFLIMFIYAILGMNWFSKVNPESGIDDIFNFKTFASSMLCLFQISTSAGWDSLLSPMLRSKESCNSSSENCHLPGIATSYFVSYIIISFLIVVNMYIAVILENFNTATEESEDPLGEDDFDIFYEVWEKFDPEATQFIKYSALSDFADALPEPLRVAKPNKYQFLVMDLPMVSEDRLHCMDILFAFTARVLGGSDGLDSMKAMMEEKFMEANPLKKLYEPIVTTTKRKEEERGAAIIQKAFRKYMMKVTKGDQGDQNDLENGPHSPLQTLCNGDLSSFGVAKGKVHCD
- Q9UQD0 SCN8A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAARLLAPPGPDSFKPFTPESLANIERRIAESKLKKPPKADGSHREDDEDSKPKPNSDLEAGKSLPFIYGDIPQGLVAVPLEDFDPYYLTQKTFVVLNRGKTLFRFSATPALYILSPFNLIRRIAIKILIHSVFSMIIMCTILTNCVFMTFSNPPDWSKNVEYTFTGIYTFESLVKIIARGFCIDGFTFLRDPWNWLDFSVIMMAYITEFVNLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCVVWPINFNESYLENGTKGFDWEEYINNKTNFYTVPGMLEPLLCGNSSDAGQCPEGYQCMKAGRNPNYGYTSFDTFSWAFLALFRLMTQDYWENLYQLTLRAAGKTYMIFFVLVIFVGSFYLVNLILAVVAMAYEEQNQATLEEAEQKEAEFKAMLEQLKKQQEEAQAAAMATSAGTVSEDAIEEEGEEGGGSPRSSSEISKLSSKSAKERRNRRKKRKQKELSEGEEKGDPEKVFKSESEDGMRRKAFRLPDNRIGRKFSIMNQSLLSIPGSPFLSRHNSKSSIFSFRGPGRFRDPGSENEFADDEHSTVEESEGRRDSLFIPIRARERRSSYSGYSGYSQGSRSSRIFPSLRRSVKRNSTVDCNGVVSLIGGPGSHIGGRLLPEATTEVEIKKKGPGSLLVSMDQLASYGRKDRINSIMSVVTNTLVEELEESQRKCPPCWYKFANTFLIWECHPYWIKLKEIVNLIVMDPFVDLAITICIVLNTLFMAMEHHPMTPQFEHVLAVGNLVFTGIFTAEMFLKLIAMDPYYYFQEGWNIFDGFIVSLSLMELSLADVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINQDCELPRWHMHDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLIVFMMVMVIGNLVVLNLFLALLLSSFSADNLAATDDDGEMNNLQISVIRIKKGVAWTKLKVHAFMQAHFKQREADEVKPLDELYEKKANCIANHTGADIHRNGDFQKNGNGTTSGIGSSVEKYIIDEDHMSFINNPNLTVRVPIAVGESDFENLNTEDVSSESDPEGSKDKLDDTSSSEGSTIDIKPEVEEVPVEQPEEYLDPDACFTEGCVQRFKCCQVNIEEGLGKSWWILRKTCFLIVEHNWFETFIIFMILLSSGALAFEDIYIEQRKTIRTILEYADKVFTYIFILEMLLKWTAYGFVKFFTNAWCWLDFLIVAVSLVSLIANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKYHYCFNETSEIRFEIEDVNNKTECEKLMEGNNTEIRWKNVKINFDNVGAGYLALLQVATFKGWMDIMYAAVDSRKPDEQPKYEDNIYMYIYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKIQGIVFDFVTQQAFDIVIMMLICLNMVTMMVETDTQSKQMENILYWINLVFVIFFTCECVLKMFALRHYYFTIGWNIFDFVVVILSIVGMFLADIIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIFSIFGMSNFAYVKHEAGIDDMFNFETFGNSMICLFQITTSAGWDGLLLPILNRPPDCSLDKEHPGSGFKGDCGNPSVGIFFFVSYIIISFLIVVNMYIAIILENFSVATEESADPLSEDDFETFYEIWEKFDPDATQFIEYCKLADFADALEHPLRVPKPNTIELIAMDLPMVSGDRIHCLDILFAFTKRVLGDSGELDILRQQMEERFVASNPSKVSYEPITTTLRRKQEEVSAVVLQRAYRGHLARRGFICKKTTSNKLENGGTHREKKESTPSTASLPSYDSVTKPEKEKQQRAEEGRRERAKRQKEVRESKC
- Q9Y5Y9 SCNAA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEFPIGSLETNNFRRFTPESLVEIEKQIAAKQGTKKAREKHREQKDQEEKPRPQLDLKACNQLPKFYGELPAELIGEPLEDLDPFYSTHRTFMVLNKGRTISRFSATRALWLFSPFNLIRRTAIKVSVHSWFSLFITVTILVNCVCMTRTDLPEKIEYVFTVIYTFEALIKILARGFCLNEFTYLRDPWNWLDFSVITLAYVGTAIDLRGISGLRTFRVLRALKTVSVIPGLKVIVGALIHSVKKLADVTILTIFCLSVFALVGLQLFKGNLKNKCVKNDMAVNETTNYSSHRKPDIYINKRGTSDPLLCGNGSDSGHCPDGYICLKTSDNPDFNYTSFDSFAWAFLSLFRLMTQDSWERLYQQTLRTSGKIYMIFFVLVIFLGSFYLVNLILAVVTMAYEEQNQATTDEIEAKEKKFQEALEMLRKEQEVLAALGIDTTSLHSHNGSPLTSKNASERRHRIKPRVSEGSTEDNKSPRSDPYNQRRMSFLGLASGKRRASHGSVFHFRSPGRDISLPEGVTDDGVFPGDHESHRGSLLLGGGAGQQGPLPRSPLPQPSNPDSRHGEDEHQPPPTSELAPGAVDVSAFDAGQKKTFLSAEYLDEPFRAQRAMSVVSIITSVLEELEESEQKCPPCLTSLSQKYLIWDCCPMWVKLKTILFGLVTDPFAELTITLCIVVNTIFMAMEHHGMSPTFEAMLQIGNIVFTIFFTAEMVFKIIAFDPYYYFQKKWNIFDCIIVTVSLLELGVAKKGSLSVLRSFRLLRVFKLAKSWPTLNTLIKIIGNSVGALGNLTIILAIIVFVFALVGKQLLGENYRNNRKNISAPHEDWPRWHMHDFFHSFLIVFRILCGEWIENMWACMEVGQKSICLILFLTVMVLGNLVVLNLFIALLLNSFSADNLTAPEDDGEVNNLQVALARIQVFGHRTKQALCSFFSRSCPFPQPKAEPELVVKLPLSSSKAENHIAANTARGSSGGLQAPRGPRDEHSDFIANPTVWVSVPIAEGESDLDDLEDDGGEDAQSFQQEVIPKGQQEQLQQVERCGDHLTPRSPGTGTSSEDLAPSLGETWKDESVPQVPAEGVDDTSSSEGSTVDCLDPEEILRKIPELADDLEEPDDCFTEGCIRHCPCCKLDTTKSPWDVGWQVRKTCYRIVEHSWFESFIIFMILLSSGSLAFEDYYLDQKPTVKALLEYTDRVFTFIFVFEMLLKWVAYGFKKYFTNAWCWLDFLIVNISLISLTAKILEYSEVAPIKALRTLRALRPLRALSRFEGMRVVVDALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFWRCINYTDGEFSLVPLSIVNNKSDCKIQNSTGSFFWVNVKVNFDNVAMGYLALLQVATFKGWMDIMYAAVDSREVNMQPKWEDNVYMYLYFVIFIIFGGFFTLNLFVGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKFQGFVFDIVTRQAFDITIMVLICLNMITMMVETDDQSEEKTKILGKINQFFVAVFTGECVMKMFALRQYYFTNGWNVFDFIVVVLSIASLIFSAILKSLQSYFSPTLFRVIRLARIGRILRLIRAAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMSSFPHVRWEAGIDDMFNFQTFANSMLCLFQITTSAGWDGLLSPILNTGPPYCDPNLPNSNGTRGDCGSPAVGIIFFTTYIIISFLIMVNMYIAVILENFNVATEESTEPLSEDDFDMFYETWEKFDPEATQFITFSALSDFADTLSGPLRIPKPNRNILIQMDLPLVPGDKIHCLDILFAFTKNVLGESGELDSLKANMEEKFMATNLSKSSYEPIATTLRWKQEDISATVIQKAYRSYVLHRSMALSNTPCVPRAEEEAASLPDEGFVAFTANENCVLPDKSETASATSFPPSYESVTRGLSDRVNMRTSSSIQNEDEATSMELIAPGP
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. Plays a key role in brain, probably by regulating the moment when neurotransmitters are released in neurons. Involved in sensory perception of mechanical pain: activation in somatosensory neurons induces pain without neurogenic inflammation and produces hypersensitivity to mechanical, but not thermal stimuli.
Phosphorylation at Ser-1516 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
The voltage-sensitive sodium channel consists of an ion conducting pore forming alpha-subunit regulated by one or more beta-1 (SCN1B), beta-2 (SCN2B), beta-3 (SCN3B) and/or beta-4 (SCN4B). Beta-1 (SCN1B) and beta-3 (SCN3B) are non-covalently associated with alpha, while beta-2 (SCN2B) and beta-4 (SCN4B) are covalently linked by disulfide bonds. Interacts with FGF13. Interacts with SCN1B. Interacts with the conotoxin GVIIJ. Interacts with the spider beta/delta-theraphotoxin-Pre1a.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
The S3b-S4 and S1-S2 loops of repeat IV are targeted by H.maculata toxins Hm1a and Hm1b, leading to inhibit fast inactivation of Nav1.1/SCN1A. Selectivity for H.maculata toxins Hm1a and Hm1b depends on S1-S2 loops of repeat IV.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.1/SCN1A subfamily.
Pore-forming subunit of a voltage-gated sodium channel complex through which Na(+) ions pass in accordance with their electrochemical gradient. Alternates between resting, activated and inactivated states. Required for normal muscle fiber excitability, normal muscle contraction and relaxation cycles, and constant muscle strength in the presence of fluctuating K(+) levels.
Phosphorylation at Ser-1328 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
Component of a voltage-sensitive sodium channel complex that consists of an ion-conducting pore-forming alpha subunit and one or more regulatory beta subunits. Interacts with SCN1B. Heterooligomer with SCN2B or SCN4B; disulfide-linked (By similarity). Interacts with the PDZ domain of the syntrophins SNTA1, SNTB1 and SNTB2 (By similarity). Interacts with the conotoxin GVIIJ.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.4/SCN4A subfamily.
This protein mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-resistant Na(+) channel isoform. This channel is responsible for the initial upstroke of the action potential. Channel inactivation is regulated by intracellular calcium levels.
Ubiquitinated by NEDD4L; which promotes its endocytosis. Does not seem to be ubiquitinated by NEDD4 or WWP2.
Phosphorylation at Ser-1503 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents (Probable). Regulated through phosphorylation by CaMK2D (By similarity).
Lacks the cysteine which covalently binds the conotoxin GVIIJ. This cysteine (position 868) is speculated in other sodium channel subunits alpha to be implied in covalent binding with the sodium channel subunit beta-2 or beta-4.
Cell membrane>Multi-pass membrane protein. Cytoplasm>Perinuclear region. Cell membrane>Sarcolemma>T-tubule.
Note: RANGRF promotes trafficking to the cell membrane.
Found in jejunal circular smooth muscle cells (at protein level). Expressed in human atrial and ventricular cardiac muscle but not in adult skeletal muscle, brain, myometrium, liver, or spleen. Isoform 4 is expressed in brain.
Interacts with the PDZ domain of the syntrophin SNTA1, SNTB1 and SNTB2 (By similarity). Interacts with NEDD4, NEDD4L, WWP2 and GPD1L. Interacts with CALM. Interacts with FGF13; the interaction is direct and FGF13 may regulate SNC5A density at membranes and function. May also interact with FGF12 and FGF14. Interacts with the spider Jingzhaotoxin-I (AC P83974, AC B1P1B7, AC B1P1B8). Interacts with ANK3.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
The IQ domain mediates association with calmodulin.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.5/SCN5A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-sensitive Na(+) channel isoform. Plays a role in pain mechanisms, especially in the development of inflammatory pain.
Phosphorylation at Ser-1490 by PKC in a highly conserved cytoplasmic loop increases peak sodium currents.
Ubiquitinated by NEDD4L; which may promote its endocytosis. Does not seem to be ubiquitinated by NEDD4.
Cell membrane>Multi-pass membrane protein. Cell projection>Neuron projection.
Note: In neurite terminals.
Expressed strongly in dorsal root ganglion, with only minor levels elsewhere in the body, smooth muscle cells, MTC cell line and C-cell carcinoma. Isoform 1 is expressed preferentially in the central and peripheral nervous system. Isoform 2 is expressed preferentially in the dorsal root ganglion.
The sodium channel complex consists of a large, channel-forming alpha subunit (SCN9A) regulated by one or more beta subunits (SCN1B, SCN2B, SCN3B and SCN4B). SCN1B and SCN3B are non-covalently associated with SCN2A. SCN2B and SCN4B are disulfide-linked to SCN2A. Interacts with NEDD4 and NEDD4L (By similarity). Interacts with the scorpion alpha-toxin CvIV4. Interacts with the conotoxin GVIIJ. Interacts with the spider huwentoxin-IV (through the extracellular loop S3-S4 of repeat II). Interacts with the spider protoxin-II (through the extracellular loop S3-S4 of repeats II and IV). Interacts with the spider beta/delta-theraphotoxin-Pre1a.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.7/SCN9A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. Implicated in the regulation of hippocampal replay occurring within sharp wave ripples (SPW-R) important for memory (By similarity).
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1506 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Sumoylated at Lys-38. Sumoylation is induced by hypoxia, increases voltage-gated sodium current and mediates the early response to acute hypoxia in neurons. Sumoylated SCN2A is located at the cell membrane.
Cell membrane>Multi-pass membrane protein.
Heterooligomer of a large alpha subunit and a smaller beta subunit. Heterooligomer with SCN2B or SCN4B; disulfide-linked. Heterooligomer with SCN1B or SCN3B; non-covalently linked. Interacts with NEDD4L. Interacts with CALM. Interacts with the conotoxin GVIIJ. Interacts with the spider beta/delta-theraphotoxin-Pre1a. Interacts with the conotoxin KIIIA. Interacts with the spider protoxin-II.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.2/SCN2A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. May contribute to the regulation of serotonin/5-hydroxytryptamine release by enterochromaffin cells (By similarity). In pancreatic endocrine cells, required for both glucagon and glucose-induced insulin secretion (By similarity).
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1501 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
Expressed in enterochromaffin cells in both colon and small bowel (at protein level).
Heterooligomer of a large alpha subunit and 2-3 smaller beta subunits. Heterooligomer with SCN2B or SCN4B; disulfide-linked. Interacts with NEDD4L. Interacts with the conotoxin GVIIJ.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.3/SCN3A subfamily.
This protein mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which sodium ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-resistant sodium channel isoform. Also involved, with the contribution of the receptor tyrosine kinase NTRK2, in rapid BDNF-evoked neuronal depolarization.
Phosphorylation at Ser-1341 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
Expressed in the dorsal root ganglia and trigeminal ganglia, olfactory bulb, hippocampus, cerebellar cortex, spinal cord, spleen, small intestine and placenta.
The voltage-resistant sodium channel consists of an ion conducting pore forming alpha-subunit regulated by one or more auxiliary subunits SCN1B, SCN2B and SCN3B.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.9/SCN11A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient.
In macrophages and melanoma cells, may participate in the control of podosome and invadopodia formation.
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1497 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein. Cell projection>Axon.
Note: Mainly localizes to the axon initial segment.
Cytoplasmic vesicle.
Note: Some vesicles are localized adjacent to melanoma invadopodia and macrophage podosomes. Does not localize to the plasma membrane.
Expressed in the hippocampus with increased expression in epileptic tissue compared to normal adjacent tissue (at protein level). Isoform 5: Expressed in non-neuronal tissues, such as monocytes/macrophages.
The voltage-sensitive sodium channel consists of an ion conducting pore forming alpha-subunit regulated by one or more beta-1 (SCN1B), beta-2 (SCN2B), beta-3 (SCN3B) and/or beta-4 (SCN4B). Beta-1 (SCN1B) and beta-3 (SCN3B) are non-covalently associated with alpha, while beta-2 (SCN2B) and beta-4 (SCN4B) are covalently linked by disulfide bonds. Interacts with NEDD4 and NEDD4L (By similarity). Interacts with FGF13. Interacts with FGF14, GBG3, GBB2 and SCN1B. Interacts with the conotoxin GVIIJ. Interacts with the conotoxin GVIIJ. Interacts with the spider beta/delta-theraphotoxin-Pre1a. Interacts with CALM1; the interaction modulates the inactivation rate of SCN8A (By similarity).
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.6/SCN8A subfamily.
Tetrodotoxin-resistant channel that mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which sodium ions may pass in accordance with their electrochemical gradient. Plays a role in neuropathic pain mechanisms.
Ubiquitinated by NEDD4L; which promotes its endocytosis.
Phosphorylation at Ser-1451 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Lacks the cysteine which covalently binds the conotoxin GVIIJ. This cysteine (position 816) is speculated in other sodium channel subunits alpha to be implied in covalent binding with the sodium channel subunit beta-2 or beta-4.
Cell membrane>Multi-pass membrane protein.
Note: It can be translocated to the cell membrane through association with S100A10.
Expressed in the dorsal root ganglia and sciatic nerve.
The channel consists of an ion conducting pore forming alpha-subunit regulated by one or more associated auxiliary subunits SCN1B, SCN2B and SCN3B; electrophysiological properties may vary depending on the type of the associated beta subunits. Found in a number of complexes with PRX, DYNLT1 and PDZD2. Interacts with proteins such as FSTL1, PRX, DYNLT1, PDZD2, S100A10 and many others (By similarity). Interacts with NEDD4 and NEDD4L.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.8/SCN10A subfamily.
研究领域
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Nervous system > Dopaminergic synapse.
· Organismal Systems > Sensory system > Taste transduction.
文献引用
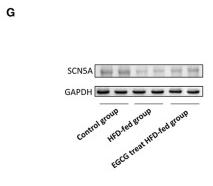
Application: WB Species: Human Sample: HepG2 cells
Application: WB Species: Rat Sample: heart
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.