产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# DF6191, RRID:AB_2838157.
展开/折叠
2 phospho D glycerate hydro lyase; 2-phospho-D-glycerate hydro-lyase; Alpha enolase; Alpha enolase like 1; Alpha-enolase; C myc promoter binding protein; C-myc promoter-binding protein; EC 4.2.1.11; eno1; ENO1L1; ENOA_HUMAN; Enolase 1 (alpha); Enolase 1 (alpha) like 1; Enolase 1; Enolase alpha; MBP 1; MBP-1; MBP1; MBPB1; MPB 1; MPB-1; MPB1; MYC promoter binding protein 1; NNE; Non neural enolase; Non-neural enolase; Phosphopyruvate hydratase; Plasminogen binding protein; Plasminogen-binding protein; PPH; Tau crystallin;
抗原和靶标
The alpha/alpha homodimer is expressed in embryo and in most adult tissues. The alpha/beta heterodimer and the beta/beta homodimer are found in striated muscle, and the alpha/gamma heterodimer and the gamma/gamma homodimer in neurons.
- P06733 ENOA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSILKIHAREIFDSRGNPTVEVDLFTSKGLFRAAVPSGASTGIYEALELRDNDKTRYMGKGVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMIEMDGTENKSKFGANAILGVSLAVCKAGAVEKGVPLYRHIADLAGNSEVILPVPAFNVINGGSHAGNKLAMQEFMILPVGAANFREAMRIGAEVYHNLKNVIKEKYGKDATNVGDEGGFAPNILENKEGLELLKTAIGKAGYTDKVVIGMDVAASEFFRSGKYDLDFKSPDDPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDDWGAWQKFTASAGIQVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLAQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSKAKFAGRNFRNPLAK
种属预测
score>80的预测可信度较高,可尝试用于WB检测。*预测模型主要基于免疫原序列比对,结果仅作参考,不作为质保凭据。
High(score>80) Medium(80>score>50) Low(score<50) No confidence
研究背景
Glycolytic enzyme the catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate. In addition to glycolysis, involved in various processes such as growth control, hypoxia tolerance and allergic responses. May also function in the intravascular and pericellular fibrinolytic system due to its ability to serve as a receptor and activator of plasminogen on the cell surface of several cell-types such as leukocytes and neurons. Stimulates immunoglobulin production.
MBP1 binds to the myc promoter and acts as a transcriptional repressor. May be a tumor suppressor.
ISGylated.
Lysine 2-hydroxyisobutyrylation (Khib) by p300/EP300 activates the phosphopyruvate hydratase activity.
Cytoplasm. Cell membrane. Cytoplasm>Myofibril>Sarcomere>M line.
Note: Can translocate to the plasma membrane in either the homodimeric (alpha/alpha) or heterodimeric (alpha/gamma) form. ENO1 is localized to the M line.
Nucleus.
The alpha/alpha homodimer is expressed in embryo and in most adult tissues. The alpha/beta heterodimer and the beta/beta homodimer are found in striated muscle, and the alpha/gamma heterodimer and the gamma/gamma homodimer in neurons.
Mammalian enolase is composed of 3 isozyme subunits, alpha, beta and gamma, which can form homodimers or heterodimers which are cell-type and development-specific. ENO1 interacts with PLG in the neuronal plasma membrane and promotes its activation. The C-terminal lysine is required for this binding. Isoform MBP-1 interacts with TRAPPC2B. Interacts with ENO4 and PGAM2 (By similarity). Interacts with CMTM6.
Belongs to the enolase family.
研究领域
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > RNA degradation.
· Metabolism > Carbohydrate metabolism > Glycolysis / Gluconeogenesis.
· Metabolism > Global and overview maps > Metabolic pathways.
· Metabolism > Global and overview maps > Carbon metabolism.
· Metabolism > Global and overview maps > Biosynthesis of amino acids.
文献引用
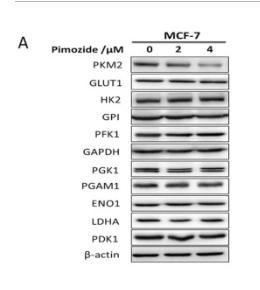
Application: WB Species: Human Sample: MCF-7 cells
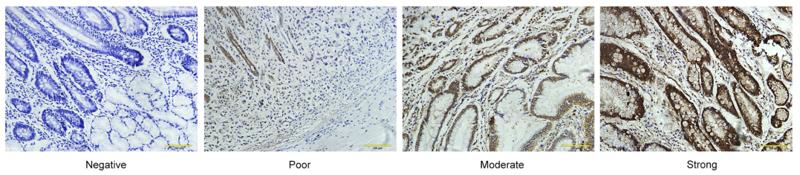
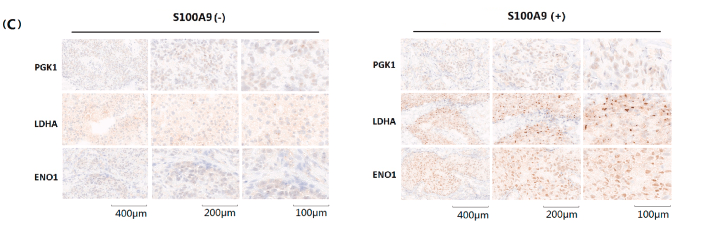
Application: IHC Species: human Sample: GC and adjacent normal tissues
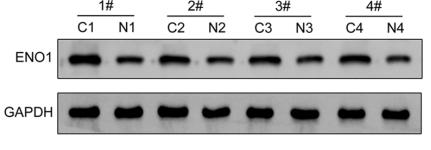
Application: WB Species: human Sample: GC tissue
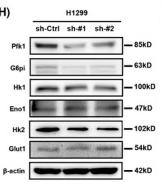
Application: WB Species: Human Sample: H1299 cells
Application: IHC Species: Human Sample: HER2+ BRCA tissues
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.