产品描述
*The optimal dilutions should be determined by the end user.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# DF6941, RRID:AB_2838900.
展开/折叠
;H3.3A; HIST1 cluster, H3E; H3 histone family, member A; H3.1; H3/l; H3F3; H3FF; H3FJ; H3FL; Histone gene cluster 1, H3 histone family, member E; histone H3.1t; Histone H3/o; FLJ92264; H 3; H3; H3 histone family, member B; H3 histone family, member C; H3 histone family, member D; H3 histone family, member F; H3 histone family, member H; H3 histone family, member I; H3 histone family, member J; H3 histone family, member K; H3 histone family, member L; H3 histone family, member T; H3 histone, family 3A; H3/A; H3/b; H3/c; H3/d; h3/f; H3/h; H3/i; H3/j; H3/k; H3/t; H31_HUMAN; H3F1K; H3F3A; H3FA; H3FB; H3FC; H3FD; H3FH; H3FI; H3FK; HIST1 cluster, H3A; HIST1 cluster, H3B; HIST1 cluster, H3C; HIST1 cluster, H3D; HIST1 cluster, H3F; HIST1 cluster, H3G; HIST1 cluster, H3H; HIST1 cluster, H3I; HIST1 cluster, H3J; HIST1H3A; HIST1H3B; HIST1H3C; HIST1H3D; HIST1H3E; HIST1H3F; HIST1H3G; HIST1H3H; HIST1H3I; HIST1H3J; HIST3H3; Histone 1, H3a; Histone 1, H3b; Histone 1, H3c; Histone 1, H3d; Histone 1, H3e; Histone 1, H3f; Histone 1, H3g; Histone 1, H3h; Histone 1, H3i; Histone 3, H3; histone cluster 1 H3 family member a; histone cluster 1 H3 family member b; histone cluster 1 H3 family member c; histone cluster 1 H3 family member d; histone cluster 1 H3 family member e; histone cluster 1 H3 family member f; histone cluster 1 H3 family member g; histone cluster 1 H3 family member h; histone cluster 1 H3 family member i; histone cluster 1 H3 family member j; Histone cluster 1, H3a; Histone cluster 1, H3b; Histone cluster 1, H3c; Histone cluster 1, H3d; Histone cluster 1, H3e; Histone cluster 1, H3f; Histone cluster 1, H3g; Histone cluster 1, H3i; Histone cluster 1, H3j; Histone gene cluster 1, H3 histone family, member A; Histone gene cluster 1, H3 histone family, member B; Histone gene cluster 1, H3 histone family, member C; Histone gene cluster 1, H3 histone family, member D; Histone gene cluster 1, H3 histone family, member F; Histone gene cluster 1, H3 histone family, member G; Histone gene cluster 1, H3 histone family, member H; Histone gene cluster 1, H3 histone family, member I; Histone gene cluster 1, H3 histone family, member J; Histone gene cluster 1, H3A; Histone gene cluster 1, H3B; Histone gene cluster 1, H3C; Histone gene cluster 1, H3D; Histone gene cluster 1, H3E; Histone gene cluster 1, H3F; Histone gene cluster 1, H3G; Histone gene cluster 1, H3H; Histone gene cluster 1, H3I; Histone gene cluster 1, H3J; Histone H 3; Histone H3.1; Histone H3.2; Histone H3.3; Histone H3/a; Histone H3/b; Histone H3/c; Histone H3/d; Histone H3/f; Histone H3/h; Histone H3/i; Histone H3/j; Histone H3/k; Histone H3/l; Histone H3/m;
抗原和靶标
- Q16695 H31T_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARTKQTARKSTGGKAPRKQLATKVARKSAPATGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLMREIAQDFKTDLRFQSSAVMALQEACESYLVGLFEDTNLCVIHAKRVTIMPKDIQLARRIRGERA
研究背景
Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
Acetylation is generally linked to gene activation. Acetylation on Lys-10 (H3K9ac) impairs methylation at Arg-9 (H3R8me2s). Acetylation on Lys-19 (H3K18ac) and Lys-24 (H3K24ac) favors methylation at Arg-18 (H3R17me). Acetylation at Lys-123 (H3K122ac) by EP300/p300 plays a central role in chromatin structure: localizes at the surface of the histone octamer and stimulates transcription, possibly by promoting nucleosome instability (By similarity).
Citrullination at Arg-9 (H3R8ci) and/or Arg-18 (H3R17ci) by PADI4 impairs methylation and represses transcription.
Asymmetric dimethylation at Arg-18 (H3R17me2a) by CARM1 is linked to gene activation. Symmetric dimethylation at Arg-9 (H3R8me2s) by PRMT5 is linked to gene repression. Asymmetric dimethylation at Arg-3 (H3R2me2a) by PRMT6 is linked to gene repression and is mutually exclusive with H3 Lys-5 methylation (H3K4me2 and H3K4me3). H3R2me2a is present at the 3' of genes regardless of their transcription state and is enriched on inactive promoters, while it is absent on active promoters (By similarity).
Methylation at Lys-5 (H3K4me), Lys-37 (H3K36me) and Lys-80 (H3K79me) are linked to gene activation. Methylation at Lys-5 (H3K4me) facilitates subsequent acetylation of H3 and H4. Methylation at Lys-80 (H3K79me) is associated with DNA double-strand break (DSB) responses and is a specific target for TP53BP1. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are linked to gene repression. Methylation at Lys-10 (H3K9me) is a specific target for HP1 proteins (CBX1, CBX3 and CBX5) and prevents subsequent phosphorylation at Ser-11 (H3S10ph) and acetylation of H3 and H4. Methylation at Lys-5 (H3K4me) and Lys-80 (H3K79me) require preliminary monoubiquitination of H2B at 'Lys-120'. Methylation at Lys-10 (H3K9me) and Lys-28 (H3K27me) are enriched in inactive X chromosome chromatin. Monomethylation at Lys-57 (H3K56me1) by EHMT2/G9A in G1 phase promotes interaction with PCNA and is required for DNA replication (By similarity).
Phosphorylated at Thr-4 (H3T3ph) by HASPIN during prophase and dephosphorylated during anaphase. Phosphorylation at Ser-11 (H3S10ph) by AURKB is crucial for chromosome condensation and cell-cycle progression during mitosis and meiosis. In addition phosphorylation at Ser-11 (H3S10ph) by RPS6KA4 and RPS6KA5 is important during interphase because it enables the transcription of genes following external stimulation, like mitogens, stress, growth factors or UV irradiation and result in the activation of genes, such as c-fos and c-jun. Phosphorylation at Ser-11 (H3S10ph), which is linked to gene activation, prevents methylation at Lys-10 (H3K9me) but facilitates acetylation of H3 and H4. Phosphorylation at Ser-11 (H3S10ph) by AURKB mediates the dissociation of HP1 proteins (CBX1, CBX3 and CBX5) from heterochromatin. Phosphorylation at Ser-11 (H3S10ph) is also an essential regulatory mechanism for neoplastic cell transformation. Phosphorylated at Ser-29 (H3S28ph) by MAP3K20 isoform 1, RPS6KA5 or AURKB during mitosis or upon ultraviolet B irradiation. Phosphorylation at Thr-7 (H3T6ph) by PRKCB is a specific tag for epigenetic transcriptional activation that prevents demethylation of Lys-5 (H3K4me) by LSD1/KDM1A. At centromeres, specifically phosphorylated at Thr-12 (H3T11ph) from prophase to early anaphase, by DAPK3 and PKN1. Phosphorylation at Thr-12 (H3T11ph) by PKN1 is a specific tag for epigenetic transcriptional activation that promotes demethylation of Lys-10 (H3K9me) by KDM4C/JMJD2C. Phosphorylation at Tyr-42 (H3Y41ph) by JAK2 promotes exclusion of CBX5 (HP1 alpha) from chromatin (By similarity).
Ubiquitinated.
Lysine deamination at Lys-5 (H3K4all) to form allysine is mediated by LOXL2. Allysine formation by LOXL2 only takes place on H3K4me3 and results in gene repression (By similarity).
Butyrylation of histones marks active promoters and competes with histone acetylation. It is present during late spermatogenesis.
Succinylation at Lys-80 (H3K79succ) by KAT2A takes place with a maximum frequency around the transcription start sites of genes. It gives a specific tag for epigenetic transcription activation. Desuccinylation at Lys-123 (H3K122succ) by SIRT7 in response to DNA damage promotes chromatin condensation and double-strand breaks (DSBs) repair.
Serine ADP-ribosylation constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage. Serine ADP-ribosylation at Ser-11 (H3S10ADPr) is mutually exclusive with phosphorylation at Ser-11 (H3S10ph) and impairs acetylation at Lys-10 (H3K9ac).
Nucleus. Chromosome.
Expressed in testicular cells.
The nucleosome is a histone octamer containing two molecules each of H2A, H2B, H3 and H4 assembled in one H3-H4 heterotetramer and two H2A-H2B heterodimers. The octamer wraps approximately 147 bp of DNA.
Belongs to the histone H3 family.
研究领域
· Human Diseases > Substance dependence > Alcoholism.
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Immune diseases > Systemic lupus erythematosus.
文献引用
Application: WB Species: Mouse Sample:
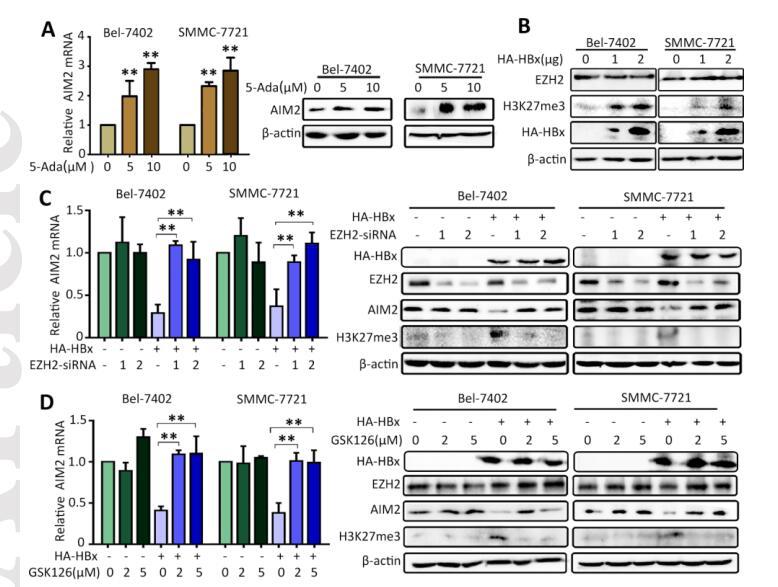
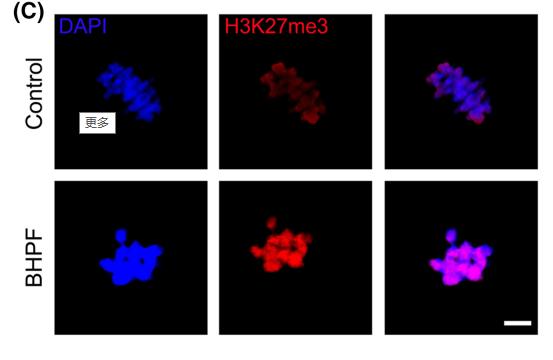
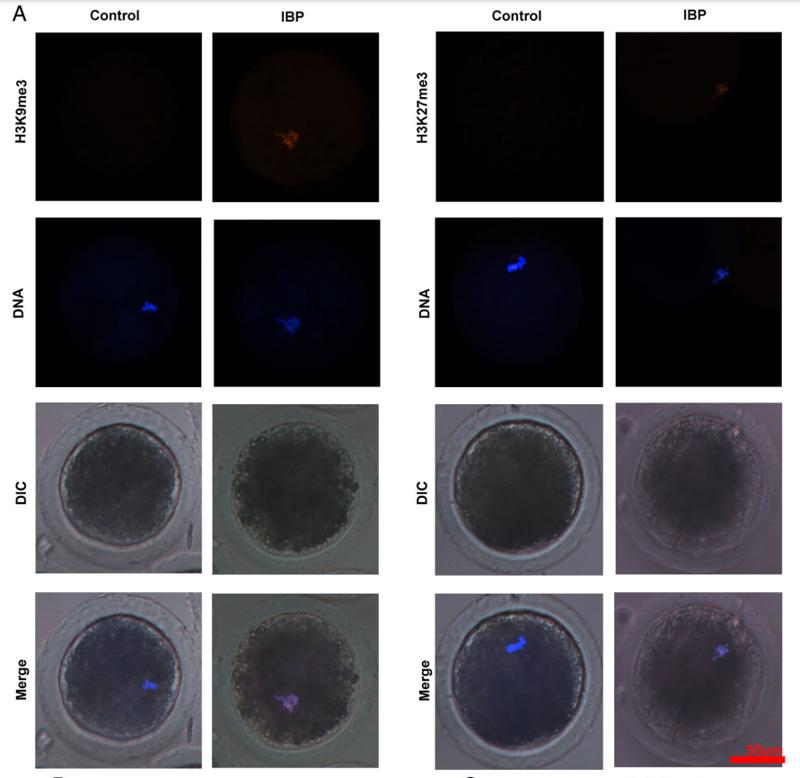
Application: IF/ICC Species: mouse Sample: oocytes
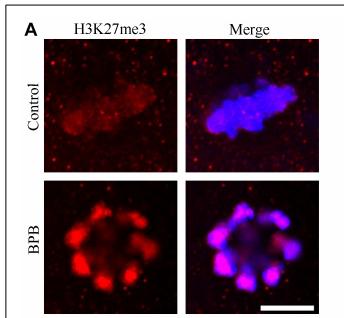
Application: IF/ICC Species: Mouse Sample:
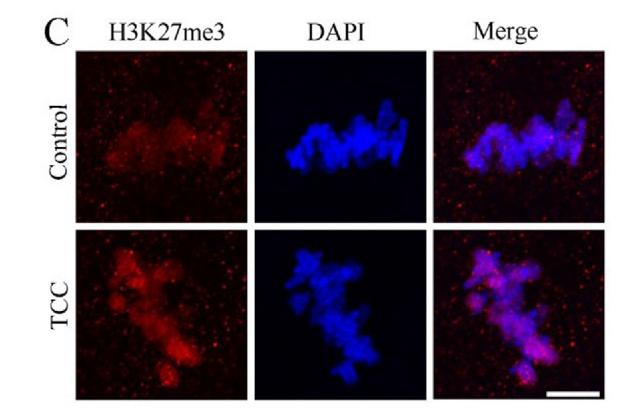
Application: IF/ICC Species: Mouse Sample:
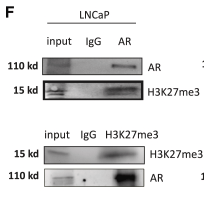
Application: WB Species: Human Sample:
Application: WB Species: Mouse Sample:
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.